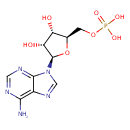
Adenosine monophosphate (ECMDB00045) (M2MDB000014)
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- A phosphate monoester + H(2)O = an alcohol + phosphate
- Gene Name:
- phoA
- Uniprot ID:
- P00634
- Molecular weight:
- 49438
Reactions
| A phosphate monoester + H(2)O = an alcohol + phosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-tryptophan + tRNA(Trp) = AMP + diphosphate + L-tryptophyl-tRNA(Trp)
- Gene Name:
- trpS
- Uniprot ID:
- P00954
- Molecular weight:
- 37438
Reactions
| ATP + L-tryptophan + tRNA(Trp) = AMP + diphosphate + L-tryptophyl-tRNA(Trp). |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of isoleucine to tRNA(Ile). As IleRS can inadvertently accommodate and process structurally similar amino acids such as valine, to avoid such errors it has two additional distinct tRNA(Ile)-dependent editing activities. One activity is designated as 'pretransfer' editing and involves the hydrolysis of activated Val-AMP. The other activity is designated 'posttransfer' editing and involves deacylation of mischarged Val-tRNA(Ile)
- Gene Name:
- ileS
- Uniprot ID:
- P00956
- Molecular weight:
- 104296
Reactions
| ATP + L-isoleucine + tRNA(Ile) = AMP + diphosphate + L-isoleucyl-tRNA(Ile). |
- General function:
- Involved in nucleotide binding
- Specific function:
- Edits incorrectly charged Ser-tRNA(Ala) and Gly- tRNA(Ala) but not incorrectly charged Ser-tRNA(Thr)
- Gene Name:
- alaS
- Uniprot ID:
- P00957
- Molecular weight:
- 96032
Reactions
| ATP + L-alanine + tRNA(Ala) = AMP + diphosphate + L-alanyl-tRNA(Ala). |
- General function:
- Involved in nucleotide binding
- Specific function:
- Is required not only for elongation of protein synthesis but also for the initiation of all mRNA translation through initiator tRNA(fMet) aminoacylation
- Gene Name:
- metG
- Uniprot ID:
- P00959
- Molecular weight:
- 76254
Reactions
| ATP + L-methionine + tRNA(Met) = AMP + diphosphate + L-methionyl-tRNA(Met). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + glycine + tRNA(Gly) = AMP + diphosphate + glycyl-tRNA(Gly)
- Gene Name:
- glyQ
- Uniprot ID:
- P00960
- Molecular weight:
- 34774
Reactions
| ATP + glycine + tRNA(Gly) = AMP + diphosphate + glycyl-tRNA(Gly). |
- General function:
- Involved in arginine-tRNA ligase activity
- Specific function:
- ATP + glycine + tRNA(Gly) = AMP + diphosphate + glycyl-tRNA(Gly)
- Gene Name:
- glyS
- Uniprot ID:
- P00961
- Molecular weight:
- 76812
Reactions
| ATP + glycine + tRNA(Gly) = AMP + diphosphate + glycyl-tRNA(Gly). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-glutamine + tRNA(Gln) = AMP + diphosphate + L-glutaminyl-tRNA(Gln)
- Gene Name:
- glnS
- Uniprot ID:
- P00962
- Molecular weight:
- 63477
Reactions
| ATP + L-glutamine + tRNA(Gln) = AMP + diphosphate + L-glutaminyl-tRNA(Gln). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-aspartate + NH(3) = AMP + diphosphate + L-asparagine
- Gene Name:
- asnA
- Uniprot ID:
- P00963
- Molecular weight:
- 36650
Reactions
| ATP + L-aspartate + NH(3) = AMP + diphosphate + L-asparagine. |
- General function:
- Involved in GMP synthase (glutamine-hydrolyzing) activity
- Specific function:
- Catalyzes the synthesis of GMP from XMP
- Gene Name:
- guaA
- Uniprot ID:
- P04079
- Molecular weight:
- 58679
Reactions
| ATP + xanthosine 5'-phosphate + L-glutamine + H(2)O = AMP + diphosphate + GMP + L-glutamate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of glutamate to tRNA(Glu) in a two-step reaction:glutamate is first activated by ATP to form Glu-AMP and then transferred to the acceptor end of tRNA(Glu)
- Gene Name:
- gltX
- Uniprot ID:
- P04805
- Molecular weight:
- 53815
Reactions
| ATP + L-glutamate + tRNA(Glu) = AMP + diphosphate + L-glutamyl-tRNA(Glu). |
- General function:
- Involved in biotin-[acetyl-CoA-carboxylase] ligase activity
- Specific function:
- BirA acts both as a biotin-operon repressor and as the enzyme that synthesizes the corepressor, acetyl-CoA:carbon-dioxide ligase. This protein also activates biotin to form biotinyl-5'- adenylate and transfers the biotin moiety to biotin-accepting proteins
- Gene Name:
- birA
- Uniprot ID:
- P06709
- Molecular weight:
- 35312
Reactions
| ATP + biotin + apo-[acetyl-CoA:carbon-dioxide ligase (ADP-forming)] = AMP + diphosphate + [acetyl-CoA:carbon-dioxide ligase (ADP-forming)]. |
- General function:
- Involved in hydrolase activity
- Specific function:
- Degradation of external UDP-glucose to uridine monophosphate and glucose-1-phosphate, which can then be used by the cell
- Gene Name:
- ushA
- Uniprot ID:
- P07024
- Molecular weight:
- 60824
Reactions
| UDP-sugar + H(2)O = UMP + alpha-D-aldose 1-phosphate. |
| A 5'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of valine to tRNA(Val). As ValRS can inadvertently accommodate and process structurally similar amino acids such as threonine, to avoid such errors, it has a "posttransfer" editing activity that hydrolyzes mischarged Thr-tRNA(Val) in a tRNA-dependent manner
- Gene Name:
- valS
- Uniprot ID:
- P07118
- Molecular weight:
- 108192
Reactions
| ATP + L-valine + tRNA(Val) = AMP + diphosphate + L-valyl-tRNA(Val). |
- General function:
- Involved in RNA binding
- Specific function:
- ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe)
- Gene Name:
- pheT
- Uniprot ID:
- P07395
- Molecular weight:
- 87377
Reactions
| ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-leucine + tRNA(Leu) = AMP + diphosphate + L-leucyl-tRNA(Leu)
- Gene Name:
- leuS
- Uniprot ID:
- P07813
- Molecular weight:
- 97233
Reactions
| ATP + L-leucine + tRNA(Leu) = AMP + diphosphate + L-leucyl-tRNA(Leu). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe)
- Gene Name:
- pheS
- Uniprot ID:
- P08312
- Molecular weight:
- 36832
Reactions
| ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe). |
- General function:
- Involved in metabolic process
- Specific function:
- Catalyzes the removal of elemental sulfur and selenium atoms from cysteine and selenocysteine to produce alanine. Functions as a sulfur delivery protein for NAD, biotin and Fe-S cluster synthesis. Transfers sulfur on 'Cys-456' of thiI in a transpersulfidation reaction. Transfers sulfur on 'Cys-19' of tusA in a transpersulfidation reaction. Functions also as a selenium delivery protein in the pathway for the biosynthesis of selenophosphate
- Gene Name:
- iscS
- Uniprot ID:
- P0A6B7
- Molecular weight:
- 45089
Reactions
| L-cysteine + acceptor = L-alanine + S-sulfanyl-acceptor. |
- General function:
- Involved in argininosuccinate synthase activity
- Specific function:
- ATP + L-citrulline + L-aspartate = AMP + diphosphate + N(omega)-(L-arginino)succinate
- Gene Name:
- argG
- Uniprot ID:
- P0A6E4
- Molecular weight:
- 49898
Reactions
| ATP + L-citrulline + L-aspartate = AMP + diphosphate + N(omega)-(L-arginino)succinate. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- ATP + D-ribose 5-phosphate = AMP + 5-phospho- alpha-D-ribose 1-diphosphate
- Gene Name:
- prs
- Uniprot ID:
- P0A717
- Molecular weight:
- 34218
Reactions
| ATP + D-ribose 5-phosphate = AMP + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in hydrolase activity
- Specific function:
- Nucleotidase with a broad substrate specificity as it can dephosphorylate various ribo- and deoxyribonucleoside 5'- monophosphates and ribonucleoside 3'-monophosphates with highest affinity to 3'-AMP. Also hydrolyzes polyphosphate (exopolyphosphatase activity) with the preference for short-chain- length substrates (P20-25). Might be involved in the regulation of dNTP and NTP pools, and in the turnover of 3'-mononucleotides produced by numerous intracellular RNases (T1, T2, and F) during the degradation of various RNAs. Also plays a significant physiological role in stress-response and is required for the survival of E.coli in stationary growth phase
- Gene Name:
- surE
- Uniprot ID:
- P0A840
- Molecular weight:
- 26900
Reactions
| A 5'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
| A 3'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
| (Polyphosphate)(n) + H(2)O = (polyphosphate)(n-1) + phosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of serine to tRNA(Ser). Is also able to aminoacylate tRNA(Sec) with serine, to form the misacylated tRNA L-seryl-tRNA(Sec), which will be further converted into selenocysteinyl-tRNA(Sec)
- Gene Name:
- serS
- Uniprot ID:
- P0A8L1
- Molecular weight:
- 48414
Reactions
| ATP + L-serine + tRNA(Ser) = AMP + diphosphate + L-seryl-tRNA(Ser). |
| ATP + L-serine + tRNA(Sec) = AMP + diphosphate + L-seryl-tRNA(Sec). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-asparagine + tRNA(Asn) = AMP + diphosphate + L-asparaginyl-tRNA(Asn)
- Gene Name:
- asnS
- Uniprot ID:
- P0A8M0
- Molecular weight:
- 52570
Reactions
| ATP + L-asparagine + tRNA(Asn) = AMP + diphosphate + L-asparaginyl-tRNA(Asn). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ThrS is also a translational repressor protein, it controls the translation of its own gene by binding to its mRNA
- Gene Name:
- thrS
- Uniprot ID:
- P0A8M3
- Molecular weight:
- 74014
Reactions
| ATP + L-threonine + tRNA(Thr) = AMP + diphosphate + L-threonyl-tRNA(Thr). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-lysine + tRNA(Lys) = AMP + diphosphate + L-lysyl-tRNA(Lys)
- Gene Name:
- lysS
- Uniprot ID:
- P0A8N3
- Molecular weight:
- 57603
Reactions
| ATP + L-lysine + tRNA(Lys) = AMP + diphosphate + L-lysyl-tRNA(Lys). |
- General function:
- Involved in nucleotide binding
- Specific function:
- Also can synthesize a number of adenyl dinucleotides (in particular AppppA). These dinucleotides have been proposed to act as modulators of the heat-shock response and stress response
- Gene Name:
- lysU
- Uniprot ID:
- P0A8N5
- Molecular weight:
- 57826
Reactions
| ATP + L-lysine + tRNA(Lys) = AMP + diphosphate + L-lysyl-tRNA(Lys). |
- General function:
- Involved in nucleotide binding
- Specific function:
- With YjeK might be involved in the post-translational modification of elongation factor P (EF-P)
- Gene Name:
- poxA
- Uniprot ID:
- P0A8N7
- Molecular weight:
- 36976
- General function:
- Involved in catalytic activity
- Specific function:
- Nucleotidase that shows high phosphatase activity toward three nucleoside 5'-monophosphates, UMP, dUMP, and dTMP, and very low activity against TDP, IMP, UDP, GMP, dGMP, AMP, dAMP, and 6- phosphogluconate. Is strictly specific to substrates with 5'- phosphates and shows no activity against nucleoside 2'- or 3'- monophosphates. Might be involved in the pyrimidine nucleotide substrate cycles
- Gene Name:
- yjjG
- Uniprot ID:
- P0A8Y1
- Molecular weight:
- 25300
Reactions
| A 5'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
- General function:
- Involved in hypoxanthine phosphoribosyltransferase activity
- Specific function:
- This enzyme acts exclusively on hypoxanthine; it does not act on guanine
- Gene Name:
- hpt
- Uniprot ID:
- P0A9M2
- Molecular weight:
- 20115
Reactions
| IMP + diphosphate = hypoxanthine + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in N6-(1,2-dicarboxyethyl)AMP AMP-lyase (fumarate-forming) activity
- Specific function:
- N(6)-(1,2-dicarboxyethyl)AMP = fumarate + AMP
- Gene Name:
- purB
- Uniprot ID:
- P0AB89
- Molecular weight:
- 51542
Reactions
| N(6)-(1,2-dicarboxyethyl)AMP = fumarate + AMP. |
| (S)-2-(5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido)succinate = fumarate + 5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide. |
- General function:
- Involved in phosphopantothenate--cysteine ligase activity
- Specific function:
- Catalyzes two steps in the biosynthesis of coenzyme A. In the first step cysteine is conjugated to 4'-phosphopantothenate to form 4-phosphopantothenoylcysteine, in the latter compound is decarboxylated to form 4'-phosphopantotheine
- Gene Name:
- coaBC
- Uniprot ID:
- P0ABQ0
- Molecular weight:
- 43438
Reactions
| N-((R)-4'-phosphopantothenoyl)-L-cysteine = pantotheine 4'-phosphate + CO(2). |
| CTP + (R)-4'-phosphopantothenate + L-cysteine = CMP + diphosphate + N-((R)-4'-phosphopantothenoyl)-L-cysteine. |
- General function:
- Involved in isochorismatase activity
- Specific function:
- Required for production of 2,3-DHB. Also serves as an aryl carrier protein and plays a role in enterobactin assembly
- Gene Name:
- entB
- Uniprot ID:
- P0ADI4
- Molecular weight:
- 32554
Reactions
| Isochorismate + H(2)O = 2,3-dihydroxy-2,3-dihydrobenzoate + pyruvate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Involved in regulation of AMP concentrations
- Gene Name:
- amn
- Uniprot ID:
- P0AE12
- Molecular weight:
- 53994
Reactions
| AMP + H(2)O = D-ribose 5-phosphate + adenine. |
- General function:
- Involved in acid phosphatase activity
- Specific function:
- Dephosphorylates several organic phosphomonoesters and catalyzes the transfer of low-energy phosphate groups from phosphomonoesters to hydroxyl groups of various organic compounds. Preferentially acts on aryl phosphoesters. Might function as a broad-spectrum dephosphorylating enzyme able to scavenge both 3'- and 5'-nucleotides and also additional organic phosphomonoesters
- Gene Name:
- aphA
- Uniprot ID:
- P0AE22
- Molecular weight:
- 26103
Reactions
| A phosphate monoester + H(2)O = an alcohol + phosphate. |
- General function:
- Involved in nucleoside-triphosphate diphosphatase activity
- Specific function:
- Specific function unknown
- Gene Name:
- mazG
- Uniprot ID:
- P0AEY3
- Molecular weight:
- 30412
Reactions
| ATP + H(2)O = AMP + diphosphate. |
- General function:
- Involved in amino acid binding
- Specific function:
- In eubacteria ppGpp (guanosine 3'-diphosphate 5-' diphosphate) is a mediator of the stringent response that coordinates a variety of cellular activities in response to changes in nutritional abundance. This enzyme catalyzes the formation of pppGpp which is then hydrolyzed to form ppGpp
- Gene Name:
- relA
- Uniprot ID:
- P0AG20
- Molecular weight:
- 83875
Reactions
| ATP + GTP = AMP + guanosine 3'-diphosphate 5'-triphosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- In eubacteria ppGpp (guanosine 3'-diphosphate 5-' diphosphate) is a mediator of the stringent response that coordinates a variety of cellular activities in response to changes in nutritional abundance. This enzyme catalyzes both the synthesis and degradation of ppGpp. The second messengers ppGpp and c-di-GMP together control biofilm formation in response to translational stress; ppGpp represses biofilm formation while c- di-GMP induces it
- Gene Name:
- spoT
- Uniprot ID:
- P0AG24
- Molecular weight:
- 79342
Reactions
| ATP + GTP = AMP + guanosine 3'-diphosphate 5'-triphosphate. |
| Guanosine 3',5'-bis(diphosphate) + H(2)O = guanosine 5'-diphosphate + diphosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of tyrosine to tRNA(Tyr) in a two-step reaction:tyrosine is first activated by ATP to form Tyr- AMP and then transferred to the acceptor end of tRNA(Tyr)
- Gene Name:
- tyrS
- Uniprot ID:
- P0AGJ9
- Molecular weight:
- 47527
Reactions
| ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + L-tyrosyl-tRNA(Tyr). |
- General function:
- Involved in catalytic activity
- Specific function:
- Activates the carboxylate group of 2,3-dihydroxy- benzoate (2,3-DHB), via ATP-dependent PPi exchange reactions, to the acyladenylate. Then, catalyzes the acylation of holo-entB with 2,3-DHB adenylate, preparing that molecule for amide bond formation with L-serine
- Gene Name:
- entE
- Uniprot ID:
- P10378
- Molecular weight:
- 59112
Reactions
| ATP + 2,3-dihydroxybenzoate = diphosphate + (2,3-dihydroxybenzoyl)adenylate. |
| (2,3-dihydroxybenzoyl)adenylate + holo-entB = adenosine 5'-monophosphate + acyl-holo-entB. |
- General function:
- Involved in ligase activity
- Specific function:
- Activates the carboxylate group of L-serine via ATP- dependent PPi exchange reactions to the aminoacyladenylate, preparing that molecule for the final stages of enterobactin synthesis. Holo-entF acts as the catalyst for the formation of the three amide and three ester bonds present in the cyclic (2,3- dihydroxybenzoyl)serine trimer enterobactin, using seryladenylate and acyl-holo-entB (acylated with 2,3-dihydroxybenzoate by entE)
- Gene Name:
- entF
- Uniprot ID:
- P11454
- Molecular weight:
- 141990
Reactions
| ATP + L-serine = diphosphate + L-serine-adenylate. |
- General function:
- Involved in phosphotransferase activity, alcohol group as acceptor
- Specific function:
- ATP + L-fuculose = ADP + L-fuculose 1- phosphate
- Gene Name:
- fucK
- Uniprot ID:
- P11553
- Molecular weight:
- 53235
Reactions
| ATP + L-fuculose = ADP + L-fuculose 1-phosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-arginine + tRNA(Arg) = AMP + diphosphate + L-arginyl-tRNA(Arg)
- Gene Name:
- argS
- Uniprot ID:
- P11875
- Molecular weight:
- 64682
Reactions
| ATP + L-arginine + tRNA(Arg) = AMP + diphosphate + L-arginyl-tRNA(Arg). |
- General function:
- Involved in DNA ligase (NAD+) activity
- Specific function:
- DNA ligase that catalyzes the formation of phosphodiester linkages between 5'-phosphoryl and 3'-hydroxyl groups in double-stranded DNA using NAD as a coenzyme and as the energy source for the reaction. It is essential for DNA replication and repair of damaged DNA
- Gene Name:
- ligA
- Uniprot ID:
- P15042
- Molecular weight:
- 73605
Reactions
| NAD(+) + (deoxyribonucleotide)(n) + (deoxyribonucleotide)(m) = AMP + beta-nicotinamide D-ribonucleotide + (deoxyribonucleotide)(n+m). |
- General function:
- Involved in catalytic activity
- Specific function:
- Synthesizes selenophosphate from selenide and ATP
- Gene Name:
- selD
- Uniprot ID:
- P16456
- Molecular weight:
- 36687
Reactions
| ATP + selenide + H(2)O = AMP + selenophosphate + phosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of proline to tRNA(Pro) in a two-step reaction:proline is first activated by ATP to form Pro- AMP and then transferred to the acceptor end of tRNA(Pro). As ProRS can inadvertently accommodate and process non-cognate amino acids such as alanine and cysteine, to avoid such errors it has two additional distinct editing activities against alanine. One activity is designated as 'pretransfer' editing and involves the tRNA(Pro)-independent hydrolysis of activated Ala-AMP. The other activity is designated 'posttransfer' editing and involves deacylation of mischarged Ala-tRNA(Pro). Misacylated Cys-tRNA(Pro) is not edited by ProRS, but instead may be edited in trans by ybaK
- Gene Name:
- proS
- Uniprot ID:
- P16659
- Molecular weight:
- 63692
Reactions
| ATP + L-proline + tRNA(Pro) = AMP + diphosphate + L-prolyl-tRNA(Pro). |
- General function:
- Involved in NAD+ synthase (glutamine-hydrolyzing) activity
- Specific function:
- Catalyzes a key step in NAD biosynthesis, transforming deamido-NAD into NAD by a two-step reaction
- Gene Name:
- nadE
- Uniprot ID:
- P18843
- Molecular weight:
- 30637
Reactions
| ATP + deamido-NAD(+) + NH(3) = AMP + diphosphate + NAD(+). |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Catalyzes the transfer of the 4'-phosphopantetheine moiety from coenzyme A to apo-domains of both entB (an ArCP domain) and entF (a PCP domain). Plays an essential role in the assembly of the enterobactin
- Gene Name:
- entD
- Uniprot ID:
- P19925
- Molecular weight:
- 23604
Reactions
| CoA + apo-EntB/F = adenosine 3',5'-bisphosphate + holo-EntB/F. |
- General function:
- Involved in cysteine-tRNA ligase activity
- Specific function:
- ATP + L-cysteine + tRNA(Cys) = AMP + diphosphate + L-cysteinyl-tRNA(Cys)
- Gene Name:
- cysS
- Uniprot ID:
- P21888
- Molecular weight:
- 52202
Reactions
| ATP + L-cysteine + tRNA(Cys) = AMP + diphosphate + L-cysteinyl-tRNA(Cys). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-aspartate + tRNA(Asp) = AMP + diphosphate + L-aspartyl-tRNA(Asp)
- Gene Name:
- aspS
- Uniprot ID:
- P21889
- Molecular weight:
- 65913
Reactions
| ATP + L-aspartate + tRNA(Asp) = AMP + diphosphate + L-aspartyl-tRNA(Asp). |
- General function:
- Involved in asparagine synthase (glutamine-hydrolyzing) activity
- Specific function:
- ATP + L-aspartate + L-glutamine + H(2)O = AMP + diphosphate + L-asparagine + L-glutamate
- Gene Name:
- asnB
- Uniprot ID:
- P22106
- Molecular weight:
- 62659
Reactions
| ATP + L-aspartate + L-glutamine + H(2)O = AMP + diphosphate + L-asparagine + L-glutamate. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Converts 3'(2')-phosphoadenosine 5'-phosphate (PAP) to AMP. May also convert adenosine 3'-phosphate 5'-phosphosulfate (PAPS) to adenosine 5'-phosphosulfate (APS). Has 10000-fold lower activity towards inositol 1,4-bisphosphate (Ins(1,4)P2)
- Gene Name:
- cysQ
- Uniprot ID:
- P22255
- Molecular weight:
- 27176
Reactions
| Adenosine 3',5'-bisphosphate + H(2)O = adenosine 5'-phosphate + phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the phosphorylation of pyruvate to phosphoenolpyruvate
- Gene Name:
- ppsA
- Uniprot ID:
- P23538
- Molecular weight:
- 87434
Reactions
| ATP + pyruvate + H(2)O = AMP + phosphoenolpyruvate + phosphate. |
- General function:
- Involved in sulfurtransferase activity
- Specific function:
- Catalyzes the 2-thiolation of uridine at the wobble position (U34) of tRNA(Lys), tRNA(Glu) and tRNA(Gln), leading to the formation of s(2)U34, the first step of tRNA-mnm(5)s(2)U34 synthesis. Sulfur is provided by iscS, via a sulfur-relay system. Binds ATP and its substrate tRNAs
- Gene Name:
- mnmA
- Uniprot ID:
- P25745
- Molecular weight:
- 40959
- General function:
- Involved in DNA ligase (NAD+) activity
- Specific function:
- Catalyzes the formation of phosphodiester linkages between 5'-phosphoryl and 3'-hydroxyl groups in double-stranded DNA using NAD as a coenzyme and as the energy source for the reaction
- Gene Name:
- ligB
- Uniprot ID:
- P25772
- Molecular weight:
- 63179
Reactions
| NAD(+) + (deoxyribonucleotide)(n) + (deoxyribonucleotide)(m) = AMP + beta-nicotinamide D-ribonucleotide + (deoxyribonucleotide)(n+m). |
- General function:
- Involved in 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase activity
- Specific function:
- ATP + 2-amino-4-hydroxy-6-hydroxymethyl-7,8- dihydropteridine = AMP + (2-amino-4-hydroxy-7,8-dihydropteridin-6- yl)methyl diphosphate
- Gene Name:
- folK
- Uniprot ID:
- P26281
- Molecular weight:
- 18079
Reactions
| ATP + 2-amino-4-hydroxy-6-hydroxymethyl-7,8-dihydropteridine = AMP + (2-amino-4-hydroxy-7,8-dihydropteridin-6-yl)methyl diphosphate. |
- General function:
- Involved in acetate-CoA ligase activity
- Specific function:
- Enables the cell to use acetate during aerobic growth to generate energy via the TCA cycle, and biosynthetic compounds via the glyoxylate shunt. Acetylates CheY, the response regulator involved in flagellar movement and chemotaxis
- Gene Name:
- acs
- Uniprot ID:
- P27550
- Molecular weight:
- 72093
Reactions
| ATP + acetate + CoA = AMP + diphosphate + acetyl-CoA. |
- General function:
- Involved in Mo-molybdopterin cofactor biosynthetic process
- Specific function:
- Involved in sulfur transfer in the conversion of molybdopterin precursor Z to molybdopterin
- Gene Name:
- moaD
- Uniprot ID:
- P30748
- Molecular weight:
- 8758
- General function:
- Involved in transferase activity, transferring acyl groups
- Specific function:
- Plays a role in lysophospholipid acylation. Transfers fatty acids to the 1-position via an enzyme-bound acyl-ACP intermediate in the presence of ATP and magnesium. Its physiological function is to regenerate phosphatidylethanolamine from 2-acyl-glycero-3-phosphoethanolamine (2-acyl-GPE) formed by transacylation reactions or degradation by phospholipase A1
- Gene Name:
- aas
- Uniprot ID:
- P31119
- Molecular weight:
- 80699
Reactions
| Acyl-[acyl-carrier-protein] + O-(2-acyl-sn-glycero-3-phospho)ethanolamine = [acyl-carrier-protein] + O-(1-beta-acyl-2-acyl-sn-glycero-3-phospho)ethanolamine. |
| ATP + an acid + [acyl-carrier-protein] = AMP + diphosphate + acyl-[acyl-carrier-protein]. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the condensation of pantoate with beta-alanine in an ATP-dependent reaction via a pantoyl-adenylate intermediate
- Gene Name:
- panC
- Uniprot ID:
- P31663
- Molecular weight:
- 31597
Reactions
| ATP + (R)-pantoate + beta-alanine = AMP + diphosphate + (R)-pantothenate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes both the ATP-dependent activation of exogenously supplied lipoate to lipoyl-AMP and the transfer of the activated lipoyl onto the lipoyl domains of lipoate-dependent enzymes. Is also able to catalyze very poorly the transfer of lipoyl and octanoyl moiety from their acyl carrier protein
- Gene Name:
- lplA
- Uniprot ID:
- P32099
- Molecular weight:
- 37926
Reactions
| ATP + lipoate = diphosphate + lipoyl-AMP. |
| Lipoyl-AMP + protein = protein N(6)-(lipoyl)lysine + AMP. |
- General function:
- Involved in hydrolase activity
- Specific function:
- NAD(+) + H(2)O = AMP + NMN
- Gene Name:
- nudC
- Uniprot ID:
- P32664
- Molecular weight:
- 29689
Reactions
| NAD(+) + H(2)O = AMP + NMN. |
- General function:
- Involved in catalytic activity
- Specific function:
- Converts 2-succinylbenzoate (OSB) to 2-succinylbenzoyl- CoA (OSB-CoA)
- Gene Name:
- menE
- Uniprot ID:
- P37353
- Molecular weight:
- 50184
Reactions
| ATP + 2-succinylbenzoate + CoA = AMP + diphosphate + 2-succinylbenzoyl-CoA. |
- General function:
- Involved in RNA processing
- Specific function:
- Catalyzes the conversion of 3'-phosphate to a 2',3'- cyclic phosphodiester at the end of RNA. The mechanism of action of the enzyme occurs in 3 steps:(A) adenylation of the enzyme by ATP; (B) the enzyme acts on RNA-N3'P to produce RNA-N3'PP5'A; (C) a non catalytic nucleophilic attack by the adjacent 2'hydroxyl on the phosphorus in the diester linkage to produce the cyclic end product. The biological role of this enzyme is unknown but it is likely to function in some aspects of cellular RNA processing
- Gene Name:
- rtcA
- Uniprot ID:
- P46849
- Molecular weight:
- 35903
Reactions
| ATP + RNA 3'-terminal-phosphate = AMP + diphosphate + RNA terminal-2',3'-cyclic-phosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-histidine + tRNA(His) = AMP + diphosphate + L-histidyl-tRNA(His)
- Gene Name:
- hisS
- Uniprot ID:
- P60906
- Molecular weight:
- 47029
Reactions
| ATP + L-histidine + tRNA(His) = AMP + diphosphate + L-histidyl-tRNA(His). |
- General function:
- Involved in ATP binding
- Specific function:
- Catalyzes the reversible transfer of the terminal phosphate group between ATP and AMP. This small ubiquitous enzyme involved in the energy metabolism and nucleotide synthesis, is essential for maintenance and cell growth
- Gene Name:
- adk
- Uniprot ID:
- P69441
- Molecular weight:
- 23586
Reactions
| ATP + AMP = 2 ADP. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the esterification, concomitant with transport, of exogenous long-chain fatty acids into metabolically active CoA thioesters for subsequent degradation or incorporation into phospholipids
- Gene Name:
- fadD
- Uniprot ID:
- P69451
- Molecular weight:
- 62332
Reactions
| ATP + a long-chain fatty acid + CoA = AMP + diphosphate + an acyl-CoA. |
- General function:
- Involved in adenine phosphoribosyltransferase activity
- Specific function:
- Catalyzes a salvage reaction resulting in the formation of AMP, that is energically less costly than de novo synthesis
- Gene Name:
- apt
- Uniprot ID:
- P69503
- Molecular weight:
- 19859
Reactions
| AMP + diphosphate = adenine + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the activation of phenylacetic acid to phenylacetyl-CoA
- Gene Name:
- paaK
- Uniprot ID:
- P76085
- Molecular weight:
- 48953
Reactions
| ATP + phenylacetate + CoA = AMP + diphosphate + phenylacetyl-CoA. |
- General function:
- Involved in phosphorus-oxygen lyase activity
- Specific function:
- In association with DosC is involved in the production and removal of the second messenger c-di-GMP in response to changing O(2) levels. Has phosphodiesterase (PDE) activity with c- di-GMP (PubMed:15995192), very poor activity on cAMP (PubMed:15995192) but not with cGMP, bis(p-nitrophenyl) phosphate or p-nitrophenyl phosphate (PubMed:11970957). Cyclic-di-GMP is a second messenger which controls cell surface-associated traits in bacteria
- Gene Name:
- dosP
- Uniprot ID:
- P76129
- Molecular weight:
- 90260
Reactions
| Cyclic di-3',5'-guanylate + H(2)O = 5'-phosphoguanylyl(3'->5')guanosine. |
- General function:
- Involved in catalytic activity
- Specific function:
- Nucleotidase that shows strict specificity toward deoxyribonucleoside 5'-monophosphates and does not dephosphorylate 5'-ribonucleotides or ribonucleoside 3'-monophosphates. Might be involved in the regulation of all dNTP pools in E.coli
- Gene Name:
- yfbR
- Uniprot ID:
- P76491
- Molecular weight:
- 22708
Reactions
| A 5'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
- General function:
- Involved in [citrate (pro-3S)-lyase] ligase activity
- Specific function:
- Acetylation of prosthetic group (2-(5''-phosphoribosyl)- 3'-dephosphocoenzyme-A) of the gamma subunit of citrate lyase
- Gene Name:
- citC
- Uniprot ID:
- P77390
- Molecular weight:
- 40077
Reactions
| ATP + acetate + [citrate (pro-3S)-lyase](thiol form) = AMP + diphosphate + [citrate (pro-3S)-lyase](acetyl form). |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the synthesis of propionyl-CoA from propionate and CoA. Also converts acetate to acetyl-CoA but with a lower specific activity
- Gene Name:
- prpE
- Uniprot ID:
- P77495
- Molecular weight:
- 69350
Reactions
| ATP + propanoate + CoA = AMP + diphosphate + propanoyl-CoA. |
- General function:
- Involved in hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
- Specific function:
- Acts on ADP-mannose and ADP-glucose as well as ADP- ribose. Prevents glycogen biosynthesis. The reaction catalyzed by this enzyme is a limiting step of the gluconeogenic process
- Gene Name:
- nudF
- Uniprot ID:
- Q93K97
- Molecular weight:
- 23667
Reactions
| ADP-ribose + H(2)O = AMP + D-ribose 5-phosphate. |
- General function:
- Involved in Mo-molybdopterin cofactor biosynthetic process
- Specific function:
- Catalyzes the insertion of molybdate into adenylated molybdopterin with the concomitant release of AMP
- Gene Name:
- moeA
- Uniprot ID:
- P12281
- Molecular weight:
- 44067
Reactions
| Adenylyl-molybdopterin + molybdate = molybdenum cofactor + AMP. |
- General function:
- Involved in hydrolase activity
- Specific function:
- Active on adenosine(5')triphospho(5')adenosine (Ap3A), ADP-ribose, NADH, adenosine(5')diphospho(5')adenosine (Ap2A)
- Gene Name:
- nudE
- Uniprot ID:
- P45799
- Molecular weight:
- 21153
Reactions
| ADP-ribose + H(2)O = AMP + D-ribose 5-phosphate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the adenylation by ATP of the carboxyl group of the C-terminal glycine of sulfur carrier protein ThiS
- Gene Name:
- thiF
- Uniprot ID:
- P30138
- Molecular weight:
- 26970
Reactions
| ATP + [ThiS] = diphosphate + adenylyl-[ThiS]. |
- General function:
- Involved in catalytic activity
- Specific function:
- Could catalyze the transfer of CoA to carnitine, generating the initial carnitinyl-CoA needed for the CaiB reaction cycle
- Gene Name:
- caiC
- Uniprot ID:
- P31552
- Molecular weight:
- 58559
- General function:
- Involved in RNA binding
- Specific function:
- Catalyzes the ATP-dependent transfer of a sulfur to tRNA to produce 4-thiouridine in position 8 of tRNAs, which functions as a near-UV photosensor. Also catalyzes the transfer of sulfur to the sulfur carrier protein ThiS, forming ThiS-thiocarboxylate. This is a step in the synthesis of thiazole, in the thiamine biosynthesis pathway. The sulfur is donated as persulfide by iscS
- Gene Name:
- thiI
- Uniprot ID:
- P77718
- Molecular weight:
- 54973
Reactions
| L-cysteine + 'activated' tRNA = L-serine + tRNA containing a thionucleotide. |
| [IscS]-SSH + [ThiS]-COAMP = [IscS]-SH + [ThiS]-COSH + AMP. |
- General function:
- Coenzyme transport and metabolism
- Specific function:
- Is the sulfur donor in the synthesis of the thiazole phosphate moiety of thiamine phosphate
- Gene Name:
- thiS
- Uniprot ID:
- O32583
- Molecular weight:
- 7311
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the radical-mediated cleavage of tyrosine to dehydroglycine and p-cresol
- Gene Name:
- thiH
- Uniprot ID:
- P30140
- Molecular weight:
- 43320
Reactions
| L-tyrosine + S-adenosyl-L-methionine + reduced acceptor = 2-iminoacetate + 4-methylphenol + 5'-deoxyadenosine + L-methionine + acceptor + 2 H(+). |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the esterification, concomitant with transport, of exogenous fatty acids into metabolically active CoA thioesters for subsequent degradation or incorporation into phospholipids. It has a low activity on medium or long chain fatty acids and is maximally active on C6 and C8 substrates
- Gene Name:
- fadK
- Uniprot ID:
- P38135
- Molecular weight:
- 60773
Reactions
| ATP + a short-chain carboxylic acid + CoA = AMP + diphosphate + an acyl-CoA. |
- General function:
- Involved in fatty acid biosynthetic process
- Specific function:
- Carrier of the growing fatty acid chain in fatty acid biosynthesis
- Gene Name:
- acpP
- Uniprot ID:
- P0A6A8
- Molecular weight:
- 8640
- General function:
- Not Available
- Specific function:
- Not Available
- Gene Name:
- tilS
- Uniprot ID:
- P52097
- Molecular weight:
- Not Available
- General function:
- Carbohydrate transport and metabolism
- Specific function:
- Not Available
- Gene Name:
- yjeF
- Uniprot ID:
- P31806
- Molecular weight:
- 54650
Reactions
| ADP + (6S)-6-beta-hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide = AMP + phosphate + NADH. |
| (R)-NADHX = (S)-NADHX. |
- General function:
- threonylcarbamoyladenosine biosynthetic process
- Specific function:
- Required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine. Is probably involved in the transfer of the threonylcarbamoyl moiety of threonylcarbamoyl-AMP (TC-AMP) to the N6 group of A37, together with TsaE and TsaB. TsaD likely plays a direct catalytic role in this reaction. May also be involved in the metabolism of glycated proteins, but does not show sialoglycoprotease activity against glycophorin A.
- Gene Name:
- tsaD
- Uniprot ID:
- P05852
- Molecular weight:
- 36008
Reactions
| L-threonylcarbamoyladenylate + adenine(37) in tRNA = AMP + N(6)-L-threonylcarbamoyladenine(37) in tRNA |
Transporters
- General function:
- Involved in transporter activity
- Specific function:
- Involved in the import of threonine and serine into the cell, with the concomitant import of a proton (symport system)
- Gene Name:
- tdcC
- Uniprot ID:
- B1XGT1
- Molecular weight:
- 48878
- General function:
- Involved in transporter activity
- Specific function:
- Non-specific porin
- Gene Name:
- ompN
- Uniprot ID:
- P77747
- Molecular weight:
- 41220
- General function:
- Involved in transporter activity
- Specific function:
- Uptake of inorganic phosphate, phosphorylated compounds, and some other negatively charged solutes
- Gene Name:
- phoE
- Uniprot ID:
- P02932
- Molecular weight:
- 38922
- General function:
- Involved in transporter activity
- Specific function:
- OmpF is a porin that forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane. It is also a receptor for the bacteriophage T2
- Gene Name:
- ompF
- Uniprot ID:
- P02931
- Molecular weight:
- 39333
- General function:
- Involved in transporter activity
- Specific function:
- Forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane
- Gene Name:
- ompC
- Uniprot ID:
- P06996
- Molecular weight:
- 40368