
Metabolites
Displaying metabolites 3626 - 3650 of 3755 in total
| Metabolite ID | Name CAS Number | IUPAC Name | SMPDB Pathways | Weight Formula | Structure |
|---|---|---|---|---|---|
| ECMDB00482 | Caprylic acid 124-07-2 | octanoic acid | 144.2114 C8H16O2 | 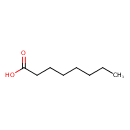 | |
| ECMDB00462 | Allantoin 97-59-6 | (2,5-dioxoimidazolidin-4-yl)urea | 158.1154 C4H6N4O3 | 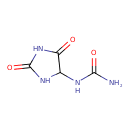 | |
| ECMDB00402 | 2-Isopropylmalic acid 3237-44-3 | (2S)-2-hydroxy-2-(propan-2-yl)butanedioic acid | 176.1672 C7H12O5 | 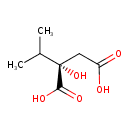 | |
| ECMDB00397 | 2-Pyrocatechuic acid 303-38-8 | 2,3-dihydroxybenzoic acid | 154.1201 C7H6O4 | 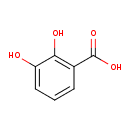 | |
| ECMDB00375 | 3-(3-Hydroxyphenyl)propanoic acid 621-54-5 | 3-(3-hydroxyphenyl)propanoic acid | 166.1739 C9H10O3 | 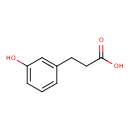 | |
| ECMDB00318 | 3,4-Dihydroxyphenylglycol 28822-73-3 | 4-(1,2-dihydroxyethyl)benzene-1,2-diol | 170.1626 C8H10O4 | 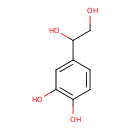 | |
| ECMDB00306 | Tyramine 51-67-2 | 4-(2-aminoethyl)phenol | 137.179 C8H11NO |  | |
| ECMDB00302 | Uridine diphosphategalactose 2956-16-3 | [({[(2S,3R,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4R,5R,6S)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy})phosphinic acid | 566.3018 C15H24N2O17P2 |  | |
| ECMDB00300 | Uracil 66-22-8 | 1,2,3,4-tetrahydropyrimidine-2,4-dione | 112.0868 C4H4N2O2 | 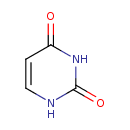 | |
| ECMDB00299 | Xanthosine 146-80-5 | 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-9H-purine-2,6-diol | purine ribonucleosides degradation PW002076 | 284.2255 C10H12N4O6 | 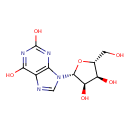 |
| ECMDB00296 | Uridine 58-96-8 | 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2,3,4-tetrahydropyrimidine-2,4-dione | 244.2014 C9H12N2O6 | 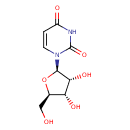 | |
| ECMDB00295 | Uridine 5'-diphosphate 58-98-0 | [({[(2R,3S,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]phosphonic acid | Pyrimidine metabolism PW000942 Starch and sucrose metabolism PW000941 Lipopolysaccharide biosynthesis PW000831 Amino sugar and nucleotide sugar metabolism III PW000895 More...Secondary Metabolites: enterobacterial common antigen biosynthesis PW000959 peptidoglycan biosynthesis I PW000906 lipopolysaccharide biosynthesis II PW001905 Secondary Metabolites: enterobacterial common antigen biosynthesis 2 PW002045 Secondary Metabolites: enterobacterial common antigen biosynthesis 3 PW002046 lipopolysaccharide biosynthesis III PW002059 peptidoglycan biosynthesis I 2 PW002062 polymyxin resistance PW002052 trehalose biosynthesis I PW002088 | 404.1612 C9H14N2O12P2 | 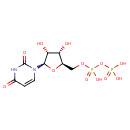 |
| ECMDB00293 | Xanthosine 5-triphosphate 6253-56-1 | ({[({[(2R,3S,4R,5R)-5-(2,6-dihydroxy-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)phosphonic acid | Collection of Reactions without pathways PW001891 | 524.1652 C10H15N4O15P3 | 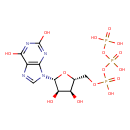 |
| ECMDB00292 | Xanthine 69-89-6 | 2,3,6,7-tetrahydro-1H-purine-2,6-dione | 152.1109 C5H4N4O2 | 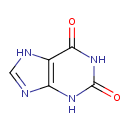 | |
| ECMDB00290 | Uridine diphosphate-N-acetylglucosamine 528-04-1 | [({[(2R,3S,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3R,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy})phosphinic acid | Lipopolysaccharide biosynthesis PW000831 Amino sugar and nucleotide sugar metabolism I PW000886 Secondary Metabolites: enterobacterial common antigen biosynthesis PW000959 peptidoglycan biosynthesis I PW000906 More...lipopolysaccharide biosynthesis II PW001905 1,6-anhydro-N-acetylmuramic acid recycling PW002064 Secondary Metabolites: enterobacterial common antigen biosynthesis 2 PW002045 Secondary Metabolites: enterobacterial common antigen biosynthesis 3 PW002046 lipopolysaccharide biosynthesis III PW002059 peptidoglycan biosynthesis I 2 PW002062 O-antigen building blocks biosynthesis PW002089 | 607.3537 C17H27N3O17P2 | 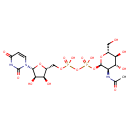 |
| ECMDB00289 | Uric acid 69-93-2 | 2,3,6,7,8,9-hexahydro-1H-purine-2,6,8-trione | adenosine nucleotides degradation PW002091 | 168.1103 C5H4N4O3 | 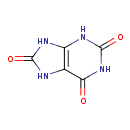 |
| ECMDB00288 | Uridine 5'-monophosphate 58-97-9 | {[(2R,3S,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}phosphonic acid | Pyrimidine metabolism PW000942 Lipopolysaccharide biosynthesis PW000831 Secondary Metabolites: enterobacterial common antigen biosynthesis PW000959 peptidoglycan biosynthesis I PW000906 More...lipopolysaccharide biosynthesis II PW001905 Secondary Metabolites: enterobacterial common antigen biosynthesis 2 PW002045 Secondary Metabolites: enterobacterial common antigen biosynthesis 3 PW002046 lipopolysaccharide biosynthesis III PW002059 peptidoglycan biosynthesis I 2 PW002062 | 324.1813 C9H13N2O9P | 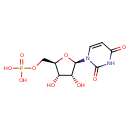 |
| ECMDB00285 | Uridine triphosphate 63-39-8 | ({[({[(2R,3S,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)phosphonic acid | Lipopolysaccharide biosynthesis PW000831 More...Amino sugar and nucleotide sugar metabolism I PW000886 Amino sugar and nucleotide sugar metabolism III PW000895 colanic acid building blocks biosynthesis PW000951 galactose degradation/Leloir Pathway PW000884 peptidoglycan biosynthesis I PW000906 lipopolysaccharide biosynthesis II PW001905 1,6-anhydro-N-acetylmuramic acid recycling PW002064 lipopolysaccharide biosynthesis III PW002059 peptidoglycan biosynthesis I 2 PW002062 O-antigen building blocks biosynthesis PW002089 | 484.1411 C9H15N2O15P3 | 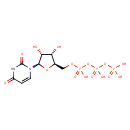 |
| ECMDB00283 | Ribose 50-69-1 | (3R,4S,5R)-5-(hydroxymethyl)oxolane-2,3,4-triol | 150.1299 C5H10O5 | 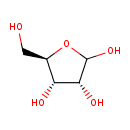 | |
| ECMDB00280 | Phosphoribosyl pyrophosphate 7540-64-9 | [({[(2R,3R,4S,5R)-3,4-dihydroxy-5-[(phosphonooxy)methyl]oxolan-2-yl]oxy}(hydroxy)phosphoryl)oxy]phosphonic acid | NAD salvage PW000830 More...PRPP Biosynthesis PW000909 Secondary Metabolites: Histidine biosynthesis PW000984 histidine biosynthesis PW000810 purine nucleotides de novo biosynthesis PW000910 purine nucleotides de novo biosynthesis 1435709748 PW000960 tryptophan metabolism II PW001916 Thiamin diphosphate biosynthesis PW002028 adenine and adenosine salvage I PW002069 adenine and adenosine salvage III PW002072 guanine and guanosine salvage PW002074 purine nucleotides de novo biosynthesis 2 PW002033 purine ribonucleosides degradation PW002076 | 390.0696 C5H13O14P3 | 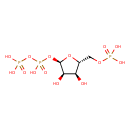 |
| ECMDB00273 | Thymidine 50-89-5 | 1-[(2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-1,2,3,4-tetrahydropyrimidine-2,4-dione | 242.2286 C10H14N2O5 | 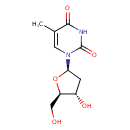 | |
| ECMDB00272 | Phosphoserine 407-41-0 | (2S)-2-amino-3-(phosphonooxy)propanoic acid | 185.0725 C3H8NO6P | 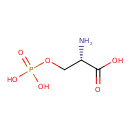 | |
| ECMDB00271 | Sarcosine 107-97-1 | 2-(methylamino)acetic acid | 89.0932 C3H7NO2 | 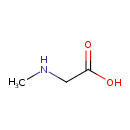 | |
| ECMDB00263 | Phosphoenolpyruvic acid 138-08-9 | 2-(phosphonooxy)prop-2-enoic acid | Lipopolysaccharide biosynthesis PW000831 Amino sugar and nucleotide sugar metabolism I PW000886 Chorismate biosynthesis PW000816 Gluconeogenesis from L-malic acid PW000819 More...Secondary Metabolites: Shikimate Pathway PW000985 fructose metabolism PW000913 glycerol metabolism PW000914 glycerol metabolism II PW000915 glycerol metabolism III (sn-glycero-3-phosphoethanolamine) PW000916 glycerol metabolism IV (glycerophosphoglycerol) PW000917 glycerol metabolism V (glycerophosphoserine) PW000918 glycolysis and pyruvate dehydrogenase PW000785 peptidoglycan biosynthesis I PW000906 superpathway of D-glucarate and D-galactarate degradation PW000795 lipopolysaccharide biosynthesis II PW001905 lipopolysaccharide biosynthesis III PW002059 peptidoglycan biosynthesis I 2 PW002062 | 168.042 C3H5O6P | 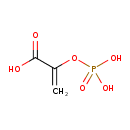 |
| ECMDB00262 | Thymine 65-71-4 | 5-methyl-1,2,3,4-tetrahydropyrimidine-2,4-dione | 126.1133 C5H6N2O2 | 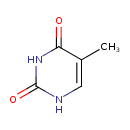 |