
Metabolites
Displaying metabolites 2426 - 2450 of 3755 in total
| Metabolite ID | Name CAS Number | IUPAC Name | SMPDB Pathways | Weight Formula | Structure |
|---|---|---|---|---|---|
| ECMDB00723 | Methylcitric acid 6061-96-7 | 2-hydroxy-1-methylpropane-1,2,3-tricarboxylic acid | Propanoate metabolism PW000940 | 206.1501 C7H10O7 | 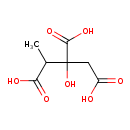 |
| ECMDB06471 | Methylisocitric acid | 1-hydroxy-1-methylpropane-1,2,3-tricarboxylic acid | Propanoate metabolism PW000940 | 206.1501 C7H10O7 | 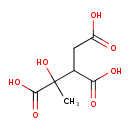 |
| ECMDB24063 | Methylmalonyl-CoA | (2S)-3-{[2-(3-{3-[({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-2-hydroxy-3-methylbutanamido}propanamido)ethyl]sulfanyl}-2-methyl-3-oxopropanoic acid | 867.607 C25H40N7O19P3S |  | |
| ECMDB20326 | Methylphosphonate 993-13-5 | methylphosphonic acid | methylphosphonate degradation I PW002065 | 96.0224 CH5O3P | 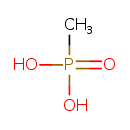 |
| ECMDB21246 | Molybdate 14259-85-9 | dihydroxydioxomolybdenumbis(ylium) | GTP degradation PW001888 | 161.95 H2MoO4 | 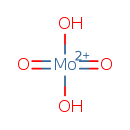 |
| ECMDB21390 | Molybdenum 7439-98-7 | molybdenum(2+) ion | 95.94 Mo |  | |
| ECMDB23236 | molybdenum cofactor 73508-07-3 | molybdenum(2+) ion ({4-hydroxy-2-imino-6,7-disulfanyl-1H,2H,5H,5aH,8H,9aH,10H-pyrano[3,2-g]pteridin-8-yl}methoxy)phosphonic acid dihydrate | GTP degradation PW001888 | 527.32 C10H18MoN5O8PS2 | 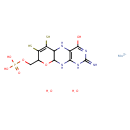 |
| ECMDB23792 | Molybdoenzyme molybdenum cofactor 73508-07-3 | {[(1R,10R,16R)-5-amino-7,13,13-trioxo-17-oxa-12,14-dithia-2,4,6,9-tetraaza-13-molybdatetracyclo[8.7.0.0³,⁸.0¹¹,¹⁵]heptadeca-3(8),5,11(15)-trien-16-yl]methoxy}phosphonic acid | 525.31 C10H16MoN5O8PS2 | 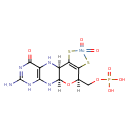 | |
| ECMDB02206 | Molybdopterin 73508-07-3 | molybdenum(2+) ion 8-[(hydrogen phosphonooxy)methyl]-2-imino-6,7-disulfanyl-1H,2H,5H,5aH,8H,9aH,10H-pyrano[3,2-g]pteridin-4-olate dihydrate | 521.27 C10H12MoN5O8PS2 | 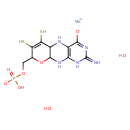 | |
| ECMDB21346 | Molybdopterin cytosine dinucleotide | [(5aR,8R,9aR)-2-amino-8-({[({[(2R,3S,4R,5R)-5-(4-amino-2-oxo-1,2-dihydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methyl phosphonato}oxy)phosphinato]oxy}methyl)-4-oxo-7-sulfanyl-1H,4H,5H,5aH,8H,9aH,10H-pyrano[3,2-g]pteridin-6-yl]sulfanide | 697.509 C19H23N8O13P2S2 | 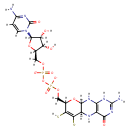 | |
| ECMDB20281 | Molybdopterin guanine dinucleotide 128007-95-4 | [({[3,4-dihydroxy-5-(6-hydroxy-2-imino-3,9-dihydro-2H-purin-9-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy][2-hydroxy-4-(4-hydroxy-2-imino-1,2,5,8-tetrahydropteridin-6-yl)-3,4-disulfanylidenebutoxy]phosphinic acid | guanylyl molybdenum cofactor biosynthesis PW002032 | 738.541 C20H24N10O13P2S2 | 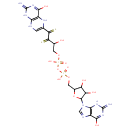 |
| ECMDB23030 | Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol | (2R)-4-{[(1S)-5-amino-1-{[(1S)-1-{[(1R)-1-carboxyethyl]-C-hydroxycarbonimidoyl}ethyl]-C-hydroxycarbonimidoyl}pentyl]-C-hydroxycarbonimidoyl}-2-{[(2S)-1-hydroxy-2-{[(2R)-1-hydroxy-2-{[(2R,3S,4R,5R,6R)-3-hydroxy-6-{[hydroxy({[hydroxy({[(2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl]oxy})phosphoryl]oxy})phosphoryl]oxy}-5-[(1-hydroxyethylidene)amino]-2-(hydroxymethyl)oxan-4-yl]oxy}propylidene]amino}propylidene]amino}butanoic acid | 1673.0374 C86H143N7O21P2 | 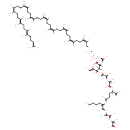 | |
| ECMDB20173 | MurAc(oyl-L-Ala-D-gamma-Glu-L-Lys-D-Ala-D-Ala)-diphospho-undecaprenol | (2R)-4-{[(1S)-5-amino-1-{[(1R)-1-{[(1R)-1-carboxyethyl]-C-hydroxycarbonimidoyl}ethyl]-C-hydroxycarbonimidoyl}pentyl]-C-hydroxycarbonimidoyl}-2-{[(2S)-1-hydroxy-2-[(1-hydroxy-2-{[(2R,3S,4R,5R,6R)-3-hydroxy-6-{[hydroxy({[hydroxy({[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl]oxy})phosphoryl]oxy})phosphoryl]oxy}-5-[(1-hydroxyethylidene)amino]-2-(hydroxymethyl)oxan-4-yl]oxy}propylidene)amino]propylidene]amino}butanoic acid | 1673.0374 C86H143N7O21P2 | 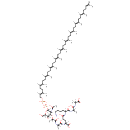 | |
| ECMDB24235 | MurNAc-6-P | (2R)-2-{[(2R,3R,4R,5S,6R)-3-acetamido-2,5-dihydroxy-6-[(phosphonatooxy)methyl]oxan-4-yl]oxy}propanoate | 370.228 C11H17NO11P | 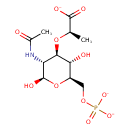 | |
| ECMDB00213 | Myo-inositol 1-phosphate 573-35-3 | {[(1R,2S,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphonic acid | 260.1358 C6H13O9P | 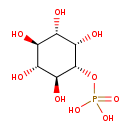 | |
| ECMDB03502 | Myo-inositol hexakisphosphate 83-86-3 | {[2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]oxy}phosphonic acid | Collection of Reactions without pathways PW001891 | 660.0353 C6H18O24P6 | 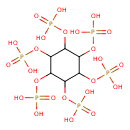 |
| ECMDB04087 | Myoinositol 87-89-8 | (1R,2R,3r,4S,5S,6s)-cyclohexane-1,2,3,4,5,6-hexol | Galactose metabolism PW000821 | 180.1559 C6H12O6 | 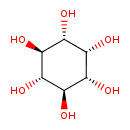 |
| ECMDB21427 | Myristic acid 544-63-8 | tetradecanoic acid | fatty acid oxidation (myristate) PW001021 | 228.3709 C14H28O2 |  |
| ECMDB24003 | N,N-Dimethylglycine 1118-68-9 | 2-(dimethylamino)acetic acid | 103.1198 C4H9NO2 | 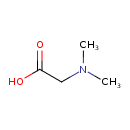 | |
| ECMDB20174 | N-(5-Phospho-D-ribosyl)anthranilate 4220-99-9 | 2-{[(3R,4S,5S)-3,4-dihydroxy-5-[hydroxy(phosphono)methyl]oxolan-2-yl]amino}benzoic acid | 349.2305 C12H16NO9P | 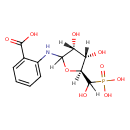 | |
| ECMDB23046 | N-Acetyl-5-glutamyl phosphate 15383-57-0 | 2-[(1-hydroxyethylidene)amino]-5-oxo-5-(phosphonooxy)pentanoic acid | 269.1458 C7H12NO8P | 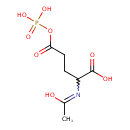 | |
| ECMDB23217 | N-Acetyl-alpha-neuraminate 21646-00-4 | (2R,4S,5R,6R)-5-acetamido-2,4-dihydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]oxane-2-carboxylic acid | 309.2699 C11H19NO9 | 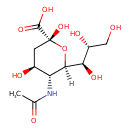 | |
| ECMDB00853 | N-Acetyl-b-D-galactosamine 14131-60-3 | N-[(2R,3R,4R,5R,6R)-2,4,5-trihydroxy-6-(hydroxymethyl)oxan-3-yl]acetamide | Galactose metabolism PW000821 | 221.2078 C8H15NO6 | 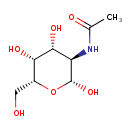 |
| ECMDB21247 | N-Acetyl-D-galactosamine 1-phosphate | (2R,3R,4R,5R,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl phosphate | 299.1718 C8H14NO9P |  | |
| ECMDB20175 | N-Acetyl-D-galactosamine 6-phosphate | N-[(3R,4R,5R,6R)-2,4,5-trihydroxy-6-[(phosphonooxy)methyl]oxan-3-yl]ethanimidic acid | Galactose metabolism PW000821 | 301.1877 C8H16NO9P |  |