
Metabolites
Displaying metabolites 401 - 425 of 3755 in total
| Metabolite ID | Name CAS Number | IUPAC Name | SMPDB Pathways | Weight Formula | Structure |
|---|---|---|---|---|---|
| ECMDB00098 | D-Xylose 58-86-6 | (3R,4S,5R)-oxane-2,3,4,5-tetrol | Starch and sucrose metabolism PW000941 | 150.1299 C5H10O5 | 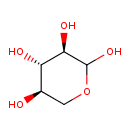 |
| ECMDB00283 | Ribose 50-69-1 | (3R,4S,5R)-5-(hydroxymethyl)oxolane-2,3,4-triol | 150.1299 C5H10O5 | 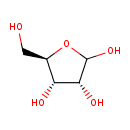 | |
| ECMDB00621 | D-Ribulose 488-84-6 | (3R,4R)-1,3,4,5-tetrahydroxypentan-2-one | D-arabinose degradation I PW002038 | 150.1299 C5H10O5 | 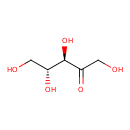 |
| ECMDB00646 | L-Arabinose 5328-37-0 | (3R,4S,5S)-oxane-2,3,4,5-tetrol | 150.1299 C5H10O5 | 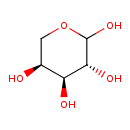 | |
| ECMDB00654 | D-Xylulose 551-84-8 | (3S,4R)-1,3,4,5-tetrahydroxypentan-2-one | 150.1299 C5H10O5 | 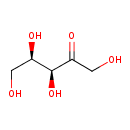 | |
| ECMDB00751 | L-Threo-2-pentulose 527-50-4 | (3R,4S)-1,3,4,5-tetrahydroxypentan-2-one | 150.1299 C5H10O5 |  | |
| ECMDB23727 | D-Arabinose | (2S,3R,4R)-2,3,4,5-tetrahydroxypentanal | Operon: arabinose activation PW001878 | 150.1299 C5H10O5 | 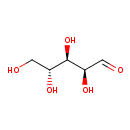 |
| ECMDB04096 | D-Lyxose 1114-34-7 | (3S,4S,5R)-oxane-2,3,4,5-tetrol | 150.1299 C5H10O5 | 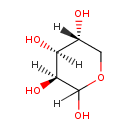 | |
| ECMDB04149 | L-Ribulose 2042-27-5 | (3S,4R)-2-(hydroxymethyl)oxolane-2,3,4-triol | L-arabinose Degradation I PW002103 | 150.1299 C5H10O5 | 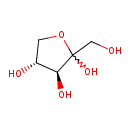 |
| ECMDB20282 | L-Ribose 24259-59-4 | (2S,3S,4S)-2,3,4,5-tetrahydroxypentanal | 150.1299 C5H10O5 | 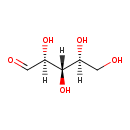 | |
| ECMDB20343 | beta-D-ribofuranose 50-69-1 | (2R,3R,4S,5R)-5-(hydroxymethyl)oxolane-2,3,4-triol | 150.1299 C5H10O5 |  | |
| ECMDB20344 | beta-D-Ribopyranose 50-69-1 | (2R,3R,4R,5R)-oxane-2,3,4,5-tetrol | Ribose Degradation PW002102 | 150.1299 C5H10O5 | 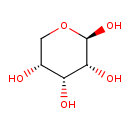 |
| ECMDB21368 | (2R,4S)-2-Methyl-2,3,3,4-tetrahydroxytetrahydrofuran | (2S,4R)-2-methyloxolane-2,3,3,4-tetrol | 150.13 C5H10O5 | 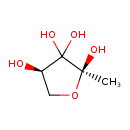 | |
| ECMDB00764 | Hydrocinnamic acid 501-52-0 | 3-phenylpropanoic acid | Phenylalanine metabolism PW000921 | 150.1745 C9H10O2 | 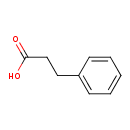 |
| ECMDB01405 | 3-Sulfinylpyruvic acid | 2-oxo-3-sulfinopropanoic acid | 151.118 C3H3O5S | 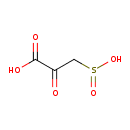 | |
| ECMDB00132 | Guanine 73-40-5 | 2-amino-6,7-dihydro-3H-purin-6-one | 151.1261 C5H5N5O | 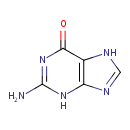 | |
| ECMDB24068 | N-Methylphenylethanolamine | 2-(methylamino)-1-phenylethan-1-ol | 151.2056 C9H13NO | 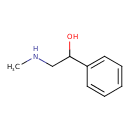 | |
| ECMDB20184 | Propanoyl phosphate 121-69-7 | (propanoyloxy)phosphonic acid | Propanoate metabolism PW000940 | 152.043 C3H5O5P | 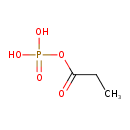 |
| ECMDB00292 | Xanthine 69-89-6 | 2,3,6,7-tetrahydro-1H-purine-2,6-dione | 152.1109 C5H4N4O2 | 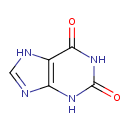 | |
| ECMDB21436 | L-Arabitol 7643-75-6 | (2S,4S)-pentane-1,2,3,4,5-pentol | 152.1458 C5H12O5 | 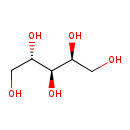 | |
| ECMDB21421 | Ribitol 488-81-3 | (2R,3s,4S)-pentane-1,2,3,4,5-pentol | 152.1458 C5H12O5 | 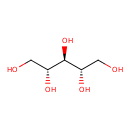 | |
| ECMDB03791 | 3,4-Dihydroxyphenylacetaldehyde 5707-55-1 | 2-(3,4-dihydroxyphenyl)acetaldehyde | 152.1473 C8H8O3 | 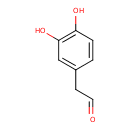 | |
| ECMDB24086 | N1-Methyl-2-pyridone-5-carboxamide | 1-methyl-6-oxo-1,6-dihydropyridine-3-carboxamide | 152.1506 C7H8N2O2 | 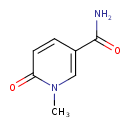 | |
| ECMDB23868 | Aromatic oxo acid | (1S,5R)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ol | 152.2334 C10H16O | 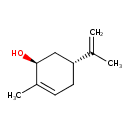 | |
| ECMDB01476 | 3-Hydroxyanthranilic acid 548-93-6 | 2-amino-3-hydroxybenzoic acid | Tryptophan metabolism PW000815 | 153.1354 C7H7NO3 | 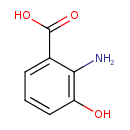 |