
Metabolites
Displaying metabolites 3576 - 3600 of 3755 in total
| Metabolite ID | Name CAS Number | IUPAC Name | SMPDB Pathways | Weight Formula | Structure |
|---|---|---|---|---|---|
| ECMDB00883 | L-Valine 72-18-4 | (2S)-2-amino-3-methylbutanoic acid | 117.1463 C5H11NO2 | 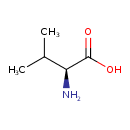 | |
| ECMDB00855 | Nicotinamide riboside 1341-23-7 | 3-carbamoyl-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1lambda5-pyridin-1-ylium | NAD salvage PW000830 | 255.2472 C11H15N2O5 | 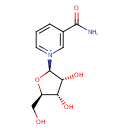 |
| ECMDB00853 | N-Acetyl-b-D-galactosamine 14131-60-3 | N-[(2R,3R,4R,5R,6R)-2,4,5-trihydroxy-6-(hydroxymethyl)oxan-3-yl]acetamide | Galactose metabolism PW000821 | 221.2078 C8H15NO6 | 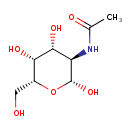 |
| ECMDB00849 | alpha-L-Rhamnose 3615-41-6 | (2R,3R,4R,5R,6S)-6-methyloxane-2,3,4,5-tetrol | 164.1565 C6H12O5 | 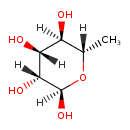 | |
| ECMDB00830 | N-Acetylneuraminic acid 131-48-6 | (2S,4S,5R,6R)-5-acetamido-2,4-dihydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]oxane-2-carboxylic acid | N-acetylneuraminate and N-acetylmannosamine and N-acetylglucosamine degradation PW002030 | 309.2699 C11H19NO9 | 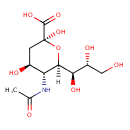 |
| ECMDB00828 | Ureidosuccinic acid 13184-27-5 | (2S)-2-(carbamoylamino)butanedioic acid | Pyrimidine metabolism PW000942 | 176.1274 C5H8N2O5 | 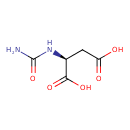 |
| ECMDB00805 | Pyrrolidonecarboxylic acid 4042-36-8 | (2R)-5-oxopyrrolidine-2-carboxylic acid | D-Glutamine and D-glutamate metabolism PW000769 | 129.114 C5H7NO3 | 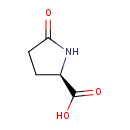 |
| ECMDB00797 | SAICAR 3031-95-6 | (2S)-2-[({5-amino-1-[(3R,4S,5S)-3,4-dihydroxy-5-[hydroxy(phosphono)methyl]oxolan-2-yl]-1H-imidazol-4-yl}(hydroxy)methylidene)amino]butanedioic acid | 454.2833 C13H19N4O12P | 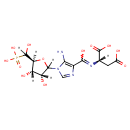 | |
| ECMDB00765 | Mannitol 69-65-8 | (2R,3R,4R,5R)-hexane-1,2,3,4,5,6-hexol | glycolysis and pyruvate dehydrogenase PW000785 | 182.1718 C6H14O6 | 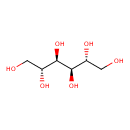 |
| ECMDB00764 | Hydrocinnamic acid 501-52-0 | 3-phenylpropanoic acid | Phenylalanine metabolism PW000921 | 150.1745 C9H10O2 | 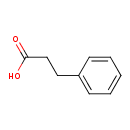 |
| ECMDB00751 | L-Threo-2-pentulose 527-50-4 | (3R,4S)-1,3,4,5-tetrahydroxypentan-2-one | 150.1299 C5H10O5 |  | |
| ECMDB00745 | Homocarnosine 3650-73-5 | (2S)-2-(4-aminobutanamido)-3-(1H-imidazol-4-yl)propanoic acid | inner membrane transport PW000786 | 240.259 C10H16N4O3 | 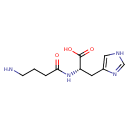 |
| ECMDB00738 | Indole 120-72-9 | 1H-indole | 117.1479 C8H7N | 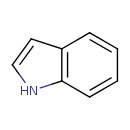 | |
| ECMDB00731 | Cysteine-S-sulfate 1637-71-4 | (2R)-2-amino-3-(sulfosulfanyl)propanoic acid | cysteine biosynthesis PW000800 | 201.221 C3H7NO5S2 | 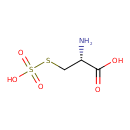 |
| ECMDB00725 | Hydroxyproline 51-35-4 | (2S,4R)-4-hydroxypyrrolidine-2-carboxylic acid | 131.1299 C5H9NO3 | 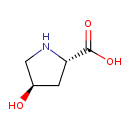 | |
| ECMDB00723 | Methylcitric acid 6061-96-7 | 2-hydroxy-1-methylpropane-1,2,3-tricarboxylic acid | Propanoate metabolism PW000940 | 206.1501 C7H10O7 | 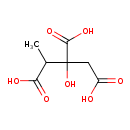 |
| ECMDB00718 | Isovaleric acid 503-74-2 | 3-methylbutanoic acid | Valine Biosynthesis PW000812 | 102.1317 C5H10O2 | 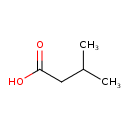 |
| ECMDB00707 | 4-Hydroxyphenylpyruvic acid 156-39-8 | 3-(4-hydroxyphenyl)-2-oxopropanoic acid | 180.1574 C9H8O4 | 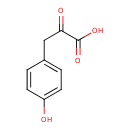 | |
| ECMDB00696 | L-Methionine 63-68-3 | (2S)-2-amino-4-(methylsulfanyl)butanoic acid | One Carbon Pool by Folate I PW001735 More...Porphyrin metabolism PW000936 S-adenosyl-L-methionine biosynthesis PW000837 methionine biosynthesis PW000814 preQ0 metabolism PW001893 tRNA Charging 2 PW000803 tRNA charging PW000799 tyrosine biosynthesis PW000806 Thiamin diphosphate biosynthesis PW002028 Thiazole Biosynthesis I PW002041 Lipoate Biosynthesis and Incorporation I PW002107 S-adenosyl-L-methionine cycle PW002080 | 149.211 C5H11NO2S | 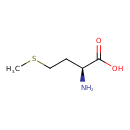 |
| ECMDB00695 | Ketoleucine 816-66-0 | 4-methyl-2-oxopentanoic acid | 130.1418 C6H10O3 | 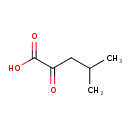 | |
| ECMDB00694 | L-2-Hydroxyglutaric acid 13095-48-2 | (2S)-2-hydroxypentanedioic acid | 148.114 C5H8O5 | 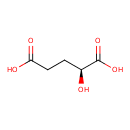 | |
| ECMDB00692 | Iron 7439-89-6 | lambda2-iron(2+) ion | 55.845 Fe |  | |
| ECMDB00687 | L-Leucine 61-90-5 | (2S)-2-amino-4-methylpentanoic acid | 131.1729 C6H13NO2 | 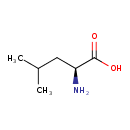 | |
| ECMDB00674 | PA(16:0/16:0) 7091-44-3 | [(2R)-2,3-bis(hexadecanoyloxy)propoxy]phosphonic acid | 648.8913 C35H69O8P | 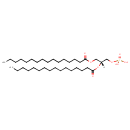 | |
| ECMDB00663 | Glucaric acid 87-73-0 | (2R,3S,4S,5S)-2,3,4,5-tetrahydroxyhexanedioic acid | superpathway of D-glucarate and D-galactarate degradation PW000795 | 210.1388 C6H10O8 | 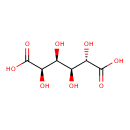 |