
Metabolites
Displaying metabolites 376 - 400 of 3755 in total
| Metabolite ID | Name CAS Number | IUPAC Name | SMPDB Pathways | Weight Formula | Structure |
|---|---|---|---|---|---|
| ECMDB00182 | L-Lysine 56-87-1 | (2S)-2,6-diaminohexanoic acid | 146.1876 C6H14N2O2 | 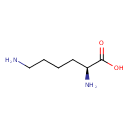 | |
| ECMDB03701 | Dimethylbenzimidazole 582-60-5 | 5,6-dimethyl-1H-1,3-benzodiazole | adenosylcobalamin salvage from cobinamide PW001884 | 146.1891 C9H10N2 | 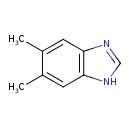 |
| ECMDB24060 | 4-Trimethylammoniobutanoic acid | (3-carboxypropyl)trimethylazanium | 146.2074 C7H16NO2 | 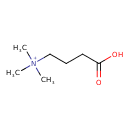 | |
| ECMDB03011 | O-Acetylserine 5147-00-2 | (2S)-3-(acetyloxy)-2-aminopropanoic acid | 147.1293 C5H9NO4 | 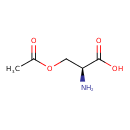 | |
| ECMDB00148 | L-Glutamate 56-86-0 | (2S)-2-aminopentanedioic acid | Phenylalanine metabolism PW000921 More...Lysine biosynthesis PW000771 Folate biosynthesis PW000908 Pantothenate and CoA biosynthesis PW000828 Tryptophan metabolism PW000815 Lipopolysaccharide biosynthesis PW000831 beta-Alanine metabolism PW000896 Propanoate metabolism PW000940 D-Glutamine and D-glutamate metabolism PW000769 Amino sugar and nucleotide sugar metabolism I PW000886 Amino sugar and nucleotide sugar metabolism III PW000895 Asparagine biosynthesis PW000813 Aspartate metabolism PW000787 Collection of Reactions without pathways PW001891 L-alanine metabolism PW000788 L-glutamate metabolism PW000789 L-glutamate metabolism II PW001886 Leucine Biosynthesis PW000811 NAD biosynthesis PW000829 NAD salvage PW000830 Porphyrin metabolism PW000936 Secondary Metabolite: Leucine biosynthesis PW000980 Secondary Metabolites: Histidine biosynthesis PW000984 Secondary Metabolites: Valine and I-leucine biosynthesis from pyruvate PW000978 Secondary Metabolites: cysteine biosynthesis from serine PW000977 Secondary Metabolites: enterobacterial common antigen biosynthesis PW000959 Valine Biosynthesis PW000812 Vitamin B6 1430936196 PW000891 arginine metabolism PW000790 cysteine biosynthesis PW000800 histidine biosynthesis PW000810 inner membrane transport PW000786 isoleucine biosynthesis PW000818 ornithine metabolism PW000791 peptidoglycan biosynthesis I PW000906 phenylalanine biosynthesis PW000807 proline metabolism PW000794 purine nucleotides de novo biosynthesis PW000910 purine nucleotides de novo biosynthesis 1435709748 PW000960 serine biosynthesis and metabolism PW000809 tRNA Charging 2 PW000803 tRNA charging PW000799 threonine biosynthesis PW000817 tyrosine biosynthesis PW000806 glutathione metabolism II PW001927 lipopolysaccharide biosynthesis II PW001905 tryptophan metabolism II PW001916 glutathione metabolism III PW002018 Thiamin diphosphate biosynthesis PW002028 4-aminobutanoate degradation I PW002068 Hydrogen Sulfide Biosynthesis I PW002066 Putrescine Degradation II PW002054 Secondary Metabolites: enterobacterial common antigen biosynthesis 2 PW002045 Secondary Metabolites: enterobacterial common antigen biosynthesis 3 PW002046 lipopolysaccharide biosynthesis III PW002059 peptidoglycan biosynthesis I 2 PW002062 polymyxin resistance PW002052 purine nucleotides de novo biosynthesis 2 PW002033 O-antigen building blocks biosynthesis PW002089 | 147.1293 C5H9NO4 | 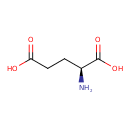 |
| ECMDB03339 | D-Glutamic acid 6893-26-1 | (2R)-2-aminopentanedioic acid | 147.1293 C5H9NO4 | 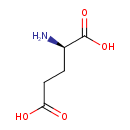 | |
| ECMDB24018 | L-Glutamic acid | (2S)-2-aminopentanedioic acid | 147.1293 C5H9NO4 | 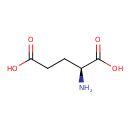 | |
| ECMDB21441 | N-Acetylserine 16354-58-8 | (2S)-2-acetamido-3-hydroxypropanoic acid | 147.1293 C5H9NO4 | 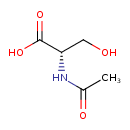 | |
| ECMDB20016 | (R)-Pantoate 470-29-1 | (2R)-2,4-dihydroxy-3,3-dimethylbutanoic acid | 147.151 C6H11O4 | 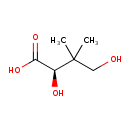 | |
| ECMDB04080 | 2-Hydroxy-3-oxosuccinate | 2-hydroxy-3-oxobutanedioic acid | glycolate and glyoxylate degradation II PW002021 | 148.071 C4H4O6 | 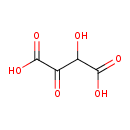 |
| ECMDB00606 | D-2-Hydroxyglutaric acid 13095-47-1 | 2-hydroxypentanedioic acid | 148.114 C5H8O5 | 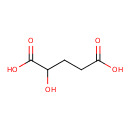 | |
| ECMDB00694 | L-2-Hydroxyglutaric acid 13095-48-2 | (2S)-2-hydroxypentanedioic acid | 148.114 C5H8O5 | 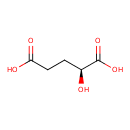 | |
| ECMDB20137 | D-Erythro-3-Methylmalate | (2R,3S)-2-hydroxy-3-methylbutanedioic acid | 148.114 C5H8O5 | 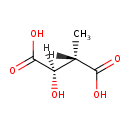 | |
| ECMDB23862 | 3-Oxoacyl-[acyl-carrier protein] | (4R)-4,5-dihydroxy-2-oxopentanoic acid | 148.114 C5H8O5 | 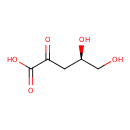 | |
| ECMDB12140 | (R) 2,3-Dihydroxy-3-methylvalerate 562-43-6 | (2R,3R)-2,3-dihydroxy-3-methylpentanoic acid | 148.1571 C6H12O4 | 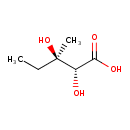 | |
| ECMDB00930 | trans-Cinnamic acid 140-10-3 | (2E)-3-phenylprop-2-enoic acid | 148.1586 C9H8O2 | 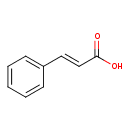 | |
| ECMDB21422 | Cinnamic acid 621-82-9 | (2Z)-3-phenylprop-2-enoic acid | 148.1586 C9H8O2 | 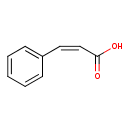 | |
| ECMDB01553 | 2-Oxo-4-methylthiobutanoic acid 583-92-6 | 4-(methylsulfanyl)-2-oxobutanoic acid | 148.18 C5H8O3S | 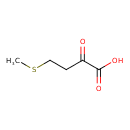 | |
| ECMDB23860 | trans-2,3-Dehydroacyl-CoA | 2-hydroxy-2-(4-hydroxyphenyl)acetonitrile | 149.1467 C8H7NO2 | 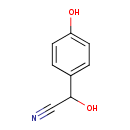 | |
| ECMDB20590 | N1-Methyladenine 5142-22-3 | 1-methyl-6,9-dihydro-1H-purin-6-imine | 149.1533 C6H7N5 | 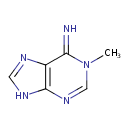 | |
| ECMDB21411 | Triethanolamine 102-71-6 | 2-[bis(2-hydroxyethyl)amino]ethan-1-ol | 149.1882 C6H15NO3 | 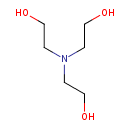 | |
| ECMDB00696 | L-Methionine 63-68-3 | (2S)-2-amino-4-(methylsulfanyl)butanoic acid | One Carbon Pool by Folate I PW001735 More...Porphyrin metabolism PW000936 S-adenosyl-L-methionine biosynthesis PW000837 methionine biosynthesis PW000814 preQ0 metabolism PW001893 tRNA Charging 2 PW000803 tRNA charging PW000799 tyrosine biosynthesis PW000806 Thiamin diphosphate biosynthesis PW002028 Thiazole Biosynthesis I PW002041 Lipoate Biosynthesis and Incorporation I PW002107 S-adenosyl-L-methionine cycle PW002080 | 149.211 C5H11NO2S | 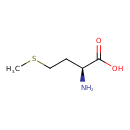 |
| ECMDB21203 | D-Methionine 348-67-4 | (2R)-2-amino-4-(methylsulfanyl)butanoic acid | 149.211 C5H11NO2S | 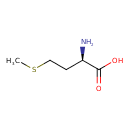 | |
| ECMDB00956 | Tartaric acid 87-69-4 | (2R,3R)-2,3-dihydroxybutanedioic acid | 150.0868 C4H6O6 | 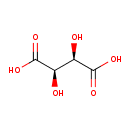 | |
| ECMDB20172 | Meso-Tartaric acid 147-73-9 | 2,3-dihydroxybutanedioic acid | 150.0868 C4H6O6 | 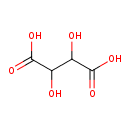 |