Carbamoylphosphate (ECMDB01096) (M2MDB000253)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 13:45:10 -0600 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 15:53:44 -0600 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Carbamoylphosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Carbamoyl phosphate is a precursor of both arginine and pyrimidine biosynthesis. It is a labile and potentially toxic intermediate. Carbamoyl phosphate is produced from carbon dioxide, ammonia, and phosphate (from ATP) by the enzyme carbamoyl phosphate synthase. -- Wikipedia | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | CH4NO5P | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 141.0199 Monoisotopic: 140.982708755 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | FFQKYPRQEYGKAF-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/CH4NO5P/c2-1(3)7-8(4,5)6/h(H2,2,3)(H2,4,5,6) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 590-55-6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (carbamoyloxy)phosphonic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | carbamoyl-phosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
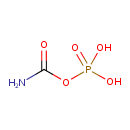
| SMILES: | NC(=O)OP(O)(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as organic phosphoric acids and derivatives. These are organic compounds containing phosphoric acid or a derivative thereof. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organic phosphoric acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Organic phosphoric acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | L-Aspartic acid + Carbamoylphosphate <> Ureidosuccinic acid + Hydrogen ion + Phosphate 2 Adenosine triphosphate + L-Glutamine + Water + Hydrogen carbonate >2 ADP + Carbamoylphosphate + L-Glutamate +2 Hydrogen ion + Phosphate Adenosine triphosphate + Carbon dioxide + Ammonium <> ADP + Carbamoylphosphate +2 Hydrogen ion Carbamoylphosphate + Ornithine + L-Ornithine <> Citrulline + Hydrogen ion + Phosphate Adenosine triphosphate + Ammonia + Carbon dioxide <> ADP + Carbamoylphosphate 2 Adenosine triphosphate + L-Glutamine + Hydrogen carbonate + Water <>2 ADP + Phosphate + L-Glutamate + Carbamoylphosphate Adenosine triphosphate + Carbamic acid <> ADP + Carbamoylphosphate Carbamoylphosphate + L-Aspartic acid <> Phosphate + Ureidosuccinic acid Carbamoylphosphate + Ornithine <> Phosphate + Citrulline L-Aspartic acid + Carbamoylphosphate > Hydrogen ion + Ureidosuccinic acid + Phosphate Ammonia + Carbon dioxide + Adenosine triphosphate < Hydrogen ion + Carbamoylphosphate + ADP Adenosine triphosphate + L-Glutamine + Hydrogen carbonate + Water > Hydrogen ion + Carbamoylphosphate + L-Glutamate + Phosphate + ADP Ornithine + Carbamoylphosphate <> Hydrogen ion + Citrulline + Phosphate Oxamate + Carbamoylphosphate < Phosphate + Oxalureate Adenosine triphosphate + Hydrogen carbonate + Ammonia > ADP + Phosphate + Carbamoylphosphate + Hydrogen ion Adenosine triphosphate + Ammonia + Carbon dioxide > ADP + Carbamoylphosphate 2 Adenosine triphosphate + L-Glutamine + Carbonic acid + Water >2 ADP + Inorganic phosphate + L-Glutamate + Carbamoylphosphate Carbamoylphosphate + Ornithine > Inorganic phosphate + Citrulline Carbamoylphosphate + L-Aspartic acid > Inorganic phosphate + Ureidosuccinic acid 2 Adenosine triphosphate + L-Glutamine + Hydrogen carbonate + Water + Ammonia + Carbamic acid + Carboxyphosphate <>2 ADP + Phosphate + L-Glutamate + Carbamoylphosphate Ornithine + Carbamoylphosphate + Ornithine > Phosphate + Hydrogen ion + Citrulline Hydrogen carbonate + Water + L-Glutamine + 2 Adenosine triphosphate >2 Adenosine diphosphate + Phosphate + L-Glutamic acid +2 Hydrogen ion + Carbamoylphosphate +2 ADP + L-Glutamate Carbamoylphosphate + L-Aspartic acid + L-Aspartic acid > Phosphate + Hydrogen ion + N-carbamoyl-L-aspartate Carbamoylphosphate + ADP + 2 Hydrogen ion > Ammonium + Adenosine triphosphate + Carbon dioxide L-Aspartic acid + Carbamoylphosphate <> Ureidosuccinic acid + Hydrogen ion + Phosphate Carbamoylphosphate + Ornithine + L-Ornithine <> Citrulline + Hydrogen ion + Phosphate More...2 Adenosine triphosphate + L-Glutamine + Water + Hydrogen carbonate >2 ADP + Carbamoylphosphate + L-Glutamate +2 Hydrogen ion + Phosphate Adenosine triphosphate + Ammonia + Carbon dioxide <> ADP + Carbamoylphosphate Adenosine triphosphate + Carbamic acid <> ADP + Carbamoylphosphate L-Aspartic acid + Carbamoylphosphate <> Ureidosuccinic acid + Hydrogen ion + Phosphate 2 Adenosine triphosphate + L-Glutamine + Water + Hydrogen carbonate >2 ADP + Carbamoylphosphate + L-Glutamate +2 Hydrogen ion + Phosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in ATP binding
- Specific function:
- 2 ATP + L-glutamine + HCO(3)(-) + H(2)O = 2 ADP + phosphate + L-glutamate + carbamoyl phosphate
- Gene Name:
- carB
- Uniprot ID:
- P00968
- Molecular weight:
- 117841
Reactions
| 2 ATP + L-glutamine + HCO(3)(-) + H(2)O = 2 ADP + phosphate + L-glutamate + carbamoyl phosphate. |
- General function:
- Involved in carboxyl- or carbamoyltransferase activity
- Specific function:
- Carbamoyl phosphate + L-ornithine = phosphate + L-citrulline
- Gene Name:
- argI
- Uniprot ID:
- P04391
- Molecular weight:
- 36907
Reactions
| Carbamoyl phosphate + L-ornithine = phosphate + L-citrulline. |
- General function:
- Involved in carboxyl- or carbamoyltransferase activity
- Specific function:
- Carbamoyl phosphate + L-ornithine = phosphate + L-citrulline
- Gene Name:
- argF
- Uniprot ID:
- P06960
- Molecular weight:
- 36827
Reactions
| Carbamoyl phosphate + L-ornithine = phosphate + L-citrulline. |
- General function:
- Involved in glutamine catabolic process
- Specific function:
- 2 ATP + L-glutamine + HCO(3)(-) + H(2)O = 2 ADP + phosphate + L-glutamate + carbamoyl phosphate
- Gene Name:
- carA
- Uniprot ID:
- P0A6F1
- Molecular weight:
- 41431
Reactions
| 2 ATP + L-glutamine + HCO(3)(-) + H(2)O = 2 ADP + phosphate + L-glutamate + carbamoyl phosphate. |
- General function:
- Involved in carboxyl- or carbamoyltransferase activity
- Specific function:
- Carbamoyl phosphate + L-aspartate = phosphate + N-carbamoyl-L-aspartate
- Gene Name:
- pyrB
- Uniprot ID:
- P0A786
- Molecular weight:
- 34427
Reactions
| Carbamoyl phosphate + L-aspartate = phosphate + N-carbamoyl-L-aspartate. |
- General function:
- Involved in 'de novo' pyrimidine base biosynthetic process
- Specific function:
- Involved in allosteric regulation of aspartate carbamoyltransferase
- Gene Name:
- pyrI
- Uniprot ID:
- P0A7F3
- Molecular weight:
- 17121
- General function:
- Involved in cellular amino acid biosynthetic process
- Specific function:
- ATP + NH(3) + CO(2) = ADP + carbamoyl phosphate
- Gene Name:
- arcC
- Uniprot ID:
- P37306
- Molecular weight:
- 31644
Reactions
| ATP + NH(3) + CO(2) = ADP + carbamoyl phosphate. |
- General function:
- Involved in cellular amino acid biosynthetic process
- Specific function:
- Specific function unknown
- Gene Name:
- yahI
- Uniprot ID:
- P77624
- Molecular weight:
- 33931
- General function:
- Involved in cellular amino acid biosynthetic process
- Specific function:
- Specific function unknown
- Gene Name:
- yqeA
- Uniprot ID:
- Q46807
- Molecular weight:
- 33071