| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:09:40 -0600 |
|---|
| Update Date | 2015-06-03 15:54:55 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
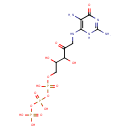
| Name: | 2,5-Diamino-6-(5'-triphosphoryl-3',4'-trihydroxy-2'-oxopentyl)-amino-4-oxopyrimidine |
|---|
| Description | 2,5-Diamino-6-(5'-triphosphoryl-3',4'-trihydroxy-2'-oxopentyl)-amino-4-oxopyrimidine is involved in folate biosynthesis. 2,5-Diamino-6-(5'-triphosphoryl-3',4'-trihydroxy-2'-oxopentyl)-amino-4-oxopyrimidine is created from 2,5-Diaminopyrimidine nucleoside triphosphate by GTP cyclohydrolase I [EC:3.5.4.16]. 2,5-Diamino-6-(5'-triphosphoryl-3',4'-trihydroxy-2'-oxopentyl)-amino-4-oxopyrimidine can be converted into 2-Amino-4-hydroxy-6-(erythro-1,2,3-trihydroxypropyl)dihydropteridine |
|---|
| Structure | |
|---|
| Synonyms: | - 1-Deoxy-1-(2,5-diamino-6-oxo-3,6-dihydro-4-pyrimidinyl)amino-5-O-(hydroxy{hydroxy(phosphonooxy)phosphoryloxy}phosphoryl)pent-2-ulose
- 5-(2,5-diamino-4-oxo-1H-pyrimidin-6-yl)amino-2,3-dihydroxy-4-oxopentoxy-hydroxyphosphoryl phosphono hydrogen phosphate
- 5-(2,5-diamino-4-oxo-1H-Pyrimidin-6-yl)amino-2,3-dihydroxy-4-oxopentoxy-hydroxyphosphoryl phosphono hydrogen phosphoric acid
|
|---|
| Chemical Formula: | C9H18N5O14P3 |
|---|
| Weight: | Average: 513.1856
Monoisotopic: 513.006309845 |
|---|
| InChI Key: | ZJYBJXKSWQPKFW-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C9H18N5O14P3/c10-5-7(13-9(11)14-8(5)18)12-1-3(15)6(17)4(16)2-26-30(22,23)28-31(24,25)27-29(19,20)21/h4,6,16-17H,1-2,10H2,(H,22,23)(H,24,25)(H2,19,20,21)(H4,11,12,13,14,18) |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | [({[({5-[(2,5-diamino-6-oxo-3,6-dihydropyrimidin-4-yl)amino]-2,3-dihydroxy-4-oxopentyl}oxy)(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl)oxy]phosphonic acid |
|---|
| Traditional IUPAC Name: | [({5-[(2,5-diamino-6-oxo-3H-pyrimidin-4-yl)amino]-2,3-dihydroxy-4-oxopentyl}oxy(hydroxy)phosphoryl)oxy(hydroxy)phosphoryl]oxyphosphonic acid |
|---|
| SMILES: | NC1=NC(=O)C(N)=C(NCC(=O)C(O)C(O)COP(O)(=O)OP(O)(=O)OP(O)(O)=O)N1 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose-5-phosphate
- Monosaccharide phosphate
- Pyrimidone
- Secondary aliphatic/aromatic amine
- Aminopyrimidine
- Monoalkyl phosphate
- Hydropyrimidine
- Phosphoric acid ester
- Beta-hydroxy ketone
- Pyrimidine
- Acyloin
- Organic phosphoric acid derivative
- Alkyl phosphate
- Alpha-hydroxy ketone
- Heteroaromatic compound
- Vinylogous amide
- 1,2-diol
- Secondary alcohol
- Ketone
- Secondary amine
- Organoheterocyclic compound
- Azacycle
- Organic nitrogen compound
- Organonitrogen compound
- Carbonyl group
- Primary amine
- Hydrocarbon derivative
- Amine
- Organic oxide
- Alcohol
- Organopnictogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -3 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0kaj-3695200000-3a55935f0cd1e0f2f9ef | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-004i-2194102000-c52d41bf51dcab1f48f4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ot-1442920000-fe5a369793012336651c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0w2c-1941200000-110e7623a57abfccc156 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udj-5910000000-57cceddb7b071bdbb766 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0492220000-7b3e1f682e2fbbb3c9a5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-057l-9560000000-056e90117fa787c6136e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-056r-9620000000-81675245a86543331ea4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000090000-0e4a8635d042911dfe6b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-06vi-2510910000-88994d38e4fc655e8499 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9200000000-15a166740d6f7d46bd0f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000190000-3bbaea893a6dbc5b8ef5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0r0a-1148930000-ef0df46fe1647dbbfe7c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000f-1943000000-1ee8cf91c2cd1830fc9b | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | HMDB06823 | | Pubchem Compound ID | 440923 | | Kegg ID | C06148 | | ChemSpider ID | 389753 | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|