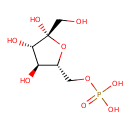
beta-D-Fructose 6-phosphate (ECMDB03971) (M2MDB000547)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:01:34 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 15:54:35 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | beta-D-Fructose 6-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Beta-D-Fructose 6 phosphate (b-F6P) is the beta-anomer of fructose-6-phosphate. There are two anomers of fructose 6 phosphate, the alpha anomer and the beta anomer. Specifically, beta-D-fructose 6-phosphate is fructose sugar phosphorylated on carbon 6. Beta-D-Fructose 6-phosphate is a substrate for Fructose-1,6-bisphosphatase, Pyruvate kinase (isozymes R/L), Hexokinase (type I), Fructose-bisphosphate aldolase A, L-lactate dehydrogenase B chain, Glyceraldehyde-3-phosphate dehydrogenase and Transaldolase. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H13O9P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 260.1358 Monoisotopic: 260.029718526 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | BGWGXPAPYGQALX-ARQDHWQXSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H13O9P/c7-2-6(10)5(9)4(8)3(15-6)1-14-16(11,12)13/h3-5,7-10H,1-2H2,(H2,11,12,13)/t3-,4-,5+,6-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(2R,3S,4S,5R)-3,4,5-trihydroxy-5-(hydroxymethyl)oxolan-2-yl]methoxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | β-D-fructofuranose 6-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC[C@@]1(O)O[C@H](COP(O)(O)=O)[C@@H](O)[C@@H]1O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pentose phosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + D-Fructose <> ADP + beta-D-Fructose 6-phosphate Mannose 6-phosphate <> beta-D-Fructose 6-phosphate Sedoheptulose 7-phosphate + D-Glyceraldehyde 3-phosphate <> D-Erythrose 4-phosphate + beta-D-Fructose 6-phosphate beta-D-Fructose 6-phosphate + D-Glyceraldehyde 3-phosphate <> D-Erythrose 4-phosphate + Xylulose 5-phosphate Sorbitol-6-phosphate + NAD <> beta-D-Fructose 6-phosphate + NADH + Hydrogen ion Glucose 6-phosphate <> beta-D-Fructose 6-phosphate beta-D-Glucose 6-phosphate <> beta-D-Fructose 6-phosphate Adenosine triphosphate + beta-D-Fructose <> ADP + beta-D-Fructose 6-phosphate Adenosine triphosphate + beta-D-Fructose 6-phosphate <> ADP + beta-D-Fructose 1,6-bisphosphate beta-D-Fructose 1,6-bisphosphate + Water <> beta-D-Fructose 6-phosphate + Phosphate D-Sedoheptulose 7-phosphate + D-Glyceraldehyde 3-phosphate + D-Sedoheptulose 7-phosphate + D-Glyceraldehyde 3-phosphate <> beta-D-Fructose 6-phosphate + D-Erythrose 4-phosphate beta-D-Glucose 6-phosphate > beta-D-Fructose 6-phosphate Sorbitol-6-phosphate + NAD <> beta-D-Fructose 6-phosphate + NADH + Hydrogen ion Sedoheptulose 7-phosphate + D-Glyceraldehyde 3-phosphate <> D-Erythrose 4-phosphate + beta-D-Fructose 6-phosphate Glucose 6-phosphate <> beta-D-Fructose 6-phosphate Sedoheptulose 7-phosphate + D-Glyceraldehyde 3-phosphate <> D-Erythrose 4-phosphate + beta-D-Fructose 6-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Find out more about how we convert literature concentrations. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in mannose-6-phosphate isomerase activity
- Specific function:
- Involved in the conversion of glucose to GDP-L-fucose, which can be converted to L-fucose, a capsular polysaccharide
- Gene Name:
- manA
- Uniprot ID:
- P00946
- Molecular weight:
- 42850
Reactions
| D-mannose 6-phosphate = D-fructose 6-phosphate. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- D-sorbitol 6-phosphate + NAD(+) = D-fructose 6-phosphate + NADH
- Gene Name:
- srlD
- Uniprot ID:
- P05707
- Molecular weight:
- 27858
Reactions
| D-sorbitol 6-phosphate + NAD(+) = D-fructose 6-phosphate + NADH. |
- General function:
- Involved in phosphotransferase activity, alcohol group as acceptor
- Specific function:
- ATP + D-fructose 6-phosphate = ADP + D- fructose 1,6-bisphosphate
- Gene Name:
- pfkB
- Uniprot ID:
- P06999
- Molecular weight:
- 32456
Reactions
| ATP + D-fructose 6-phosphate = ADP + D-fructose 1,6-bisphosphate. |
- General function:
- Involved in oxidation-reduction process
- Specific function:
- D-mannitol 1-phosphate + NAD(+) = D-fructose 6-phosphate + NADH
- Gene Name:
- mtlD
- Uniprot ID:
- P09424
- Molecular weight:
- 41139
Reactions
| D-mannitol 1-phosphate + NAD(+) = D-fructose 6-phosphate + NADH. |
- General function:
- Involved in glucose-6-phosphate isomerase activity
- Specific function:
- D-glucose 6-phosphate = D-fructose 6- phosphate
- Gene Name:
- pgi
- Uniprot ID:
- P0A6T1
- Molecular weight:
- 61529
Reactions
| D-glucose 6-phosphate = D-fructose 6-phosphate. |
- General function:
- Involved in ATP binding
- Specific function:
- ATP + D-fructose 6-phosphate = ADP + D- fructose 1,6-bisphosphate
- Gene Name:
- pfkA
- Uniprot ID:
- P0A796
- Molecular weight:
- 34842
Reactions
| ATP + D-fructose 6-phosphate = ADP + D-fructose 1,6-bisphosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Transaldolase is important for the balance of metabolites in the pentose-phosphate pathway
- Gene Name:
- talA
- Uniprot ID:
- P0A867
- Molecular weight:
- 35659
Reactions
| Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-erythrose 4-phosphate + D-fructose 6-phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Transaldolase is important for the balance of metabolites in the pentose-phosphate pathway
- Gene Name:
- talB
- Uniprot ID:
- P0A870
- Molecular weight:
- 35219
Reactions
| Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-erythrose 4-phosphate + D-fructose 6-phosphate. |
- General function:
- Involved in phosphoric ester hydrolase activity
- Specific function:
- D-fructose 1,6-bisphosphate + H(2)O = D- fructose 6-phosphate + phosphate
- Gene Name:
- fbp
- Uniprot ID:
- P0A993
- Molecular weight:
- 36834
Reactions
| D-fructose 1,6-bisphosphate + H(2)O = D-fructose 6-phosphate + phosphate. |
- General function:
- Involved in glycerol metabolic process
- Specific function:
- D-fructose 1,6-bisphosphate + H(2)O = D- fructose 6-phosphate + phosphate
- Gene Name:
- glpX
- Uniprot ID:
- P0A9C9
- Molecular weight:
- 35852
Reactions
| D-fructose 1,6-bisphosphate + H(2)O = D-fructose 6-phosphate + phosphate. |
- General function:
- Transcription
- Specific function:
- Catalyzes the phosphorylation of fructose to fructose-6- P. Has also low level glucokinase activity in vitro. Is not able to phosphorylate D-ribose, D-mannitol, D-sorbitol, inositol, and L-threonine
- Gene Name:
- mak
- Uniprot ID:
- P23917
- Molecular weight:
- 32500
Reactions
| ATP + D-fructose = ADP + D-fructose 6-phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-ribose 5-phosphate + D-xylulose 5-phosphate
- Gene Name:
- tktA
- Uniprot ID:
- P27302
- Molecular weight:
- 72211
Reactions
| Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-ribose 5-phosphate + D-xylulose 5-phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Specific function unknown
- Gene Name:
- alsE
- Uniprot ID:
- P32719
- Molecular weight:
- 26109
- General function:
- Involved in catalytic activity
- Specific function:
- Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-ribose 5-phosphate + D-xylulose 5-phosphate
- Gene Name:
- tktB
- Uniprot ID:
- P33570
- Molecular weight:
- 73042
Reactions
| Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-ribose 5-phosphate + D-xylulose 5-phosphate. |