| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 13:51:21 -0600 |
|---|
| Update Date | 2015-10-02 02:25:47 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
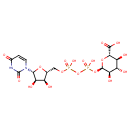
| Name: | UDP-Glucuronic acid |
|---|
| Description | UDP-Glucuronic acid is an intermediate in polymyxin resistance in E. coli. It is a substrate for UDP-GlcA C-4"-decarboxylase (gene name: arnA) which catalyzes the chemical reaction UDP-D-glucuronate + NAD+ -> UDP-beta-L-threo-pentapyranos-4-ulose + CO2 + NADH. In UDP-alpha-D-glucuronate biosynthesis (from UDP-glucose), UDP-glucose 6-dehydrogenase converts UDP-alpha-D-glucose to UDP-Glucuronic acid (EcoCyc compound: UDP-GLUCURONATE). |
|---|
| Structure | |
|---|
| Synonyms: | - UDP-D-Glucuronate
- UDP-D-Glucuronic acid
- UDP-Glucuronate
- UDP-Glucuronic acid
- UDPglucuronate
- UDPglucuronic acid
- Uridine diphosphate glucuronate
- Uridine diphosphate glucuronic acid
- Uridine diphosphoric acid glucuronic acid
|
|---|
| Chemical Formula: | C15H22N2O18P2 |
|---|
| Weight: | Average: 580.2853
Monoisotopic: 580.034284934 |
|---|
| InChI Key: | HDYANYHVCAPMJV-LXQIFKJMSA-N |
|---|
| InChI: | InChI=1S/C15H22N2O18P2/c18-5-1-2-17(15(26)16-5)12-9(22)6(19)4(32-12)3-31-36(27,28)35-37(29,30)34-14-10(23)7(20)8(21)11(33-14)13(24)25/h1-2,4,6-12,14,19-23H,3H2,(H,24,25)(H,27,28)(H,29,30)(H,16,18,26)/t4-,6-,7+,8+,9-,10-,11+,12-,14-/m1/s1 |
|---|
| CAS number: | 2616-64-0 |
|---|
| IUPAC Name: | (2S,3S,4S,5R,6R)-6-({[({[(2R,3S,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-3,4,5-trihydroxyoxane-2-carboxylic acid |
|---|
| Traditional IUPAC Name: | udp-α-D-glucuronic acid |
|---|
| SMILES: | O[C@@H]1[C@@H](COP(O)(=O)OP(O)(=O)O[C@H]2O[C@@H]([C@@H](O)[C@H](O)[C@H]2O)C(O)=O)O[C@H]([C@@H]1O)N1C=CC(=O)NC1=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as gamma butyrolactones. Gamma butyrolactones are compounds containing a gamma butyrolactone moiety, which consists of an aliphatic five-member ring with four carbon atoms, one oxygen atom, and bears a ketone group on the carbon adjacent to the oxygen atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Lactones |
|---|
| Sub Class | Gamma butyrolactones |
|---|
| Direct Parent | Gamma butyrolactones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Gamma butyrolactone
- Tetrahydrofuran
- Secondary alcohol
- Carboxylic acid ester
- 1,2-diol
- Oxacycle
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -3 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | Amino sugar and nucleotide sugar metabolism III | PW000895 |  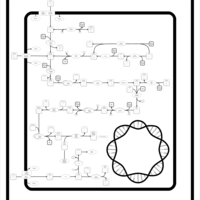 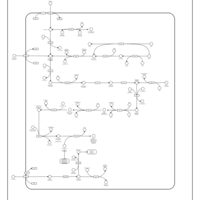 | | Starch and sucrose metabolism | PW000941 | 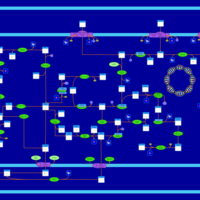 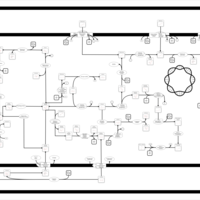 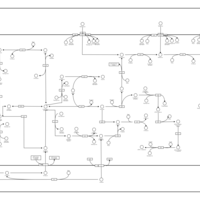 | | colanic acid building blocks biosynthesis | PW000951 |  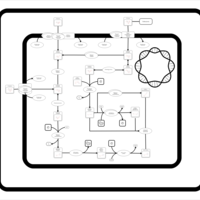 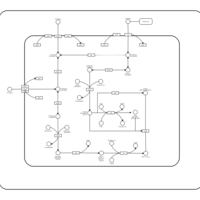 | | polymyxin resistance | PW002052 | 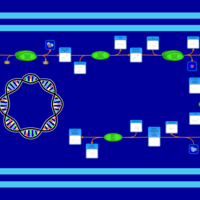 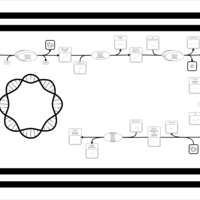 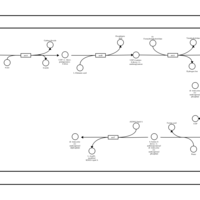 |
|
|---|
| KEGG Pathways: | - Amino sugar and nucleotide sugar metabolism ec00520
- Ascorbate and aldarate metabolism ec00053
- Metabolic pathways eco01100
- Pentose and glucuronate interconversions ec00040
- Starch and sucrose metabolism ec00500
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0w29-3322930000-0ce18416678ea80bec67 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-004r-4862914000-b91973e084ab99d92364 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Uridine diphosphate glucuronic acid,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0fb9-0135690000-0ff1e6a4900ecdc6a915 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-004i-0000090000-1cdeb28950f03fda28ea | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0r29-5934300000-e5fcfe66852c383b82d4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000090000-bc00d530c5ce60dfe672 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-3922200000-353e174f91c8b182064c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0901110000-d434fad6ebdd7797c473 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-3911000000-b27e8f43f987aadd35fd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-4900000000-6c8ee9c73e164f9ccf76 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0imu-4601190000-7a0e82462b874f18b2c4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01ox-9817020000-a72141b1bee762a83bc3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0bvl-4901000000-5d52b1b8f03063646a7f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0001090000-c773521e9e340d5e6564 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-055e-9424840000-deea5ee6888efff36f31 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a5a-3936510000-b41af0ea55c95b9da1e1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-1700490000-7f9159b24d34e9358740 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-4921760000-cd1fe2a62f343b8eb5ba | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0209-6940100000-aa916ad8a0ed15e6eb2a | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Alkharfy KM, Frye RF: High-performance liquid chromatographic assay for acetaminophen glucuronide in human liver microsomes. J Chromatogr B Biomed Sci Appl. 2001 Apr 5;753(2):303-8. Pubmed: 11334344
- Cappiello M, Giuliani L, Pacifici GM: Distribution of UDP-glucuronosyltransferase and its endogenous substrate uridine 5'-diphosphoglucuronic acid in human tissues. Eur J Clin Pharmacol. 1991;41(4):345-50. Pubmed: 1804651
- Cappiello M, Giuliani L, Rane A, Pacifici GM: Uridine 5'-diphosphoglucuronic acid (UDPGLcUA) in the human fetal liver, kidney and placenta. Eur J Drug Metab Pharmacokinet. 2000 Jul-Dec;25(3-4):161-3. Pubmed: 11420884
- Clinical Guide to Laboratory Tests, 2nd Ed. Norbert W. Tietz 1990
- Fondeur-Gelinotte M, Lattard V, Oriol R, Mollicone R, Jacquinet JC, Mulliert G, Gulberti S, Netter P, Magdalou J, Ouzzine M, Fournel-Gigleux S: Phylogenetic and mutational analyses reveal key residues for UDP-glucuronic acid binding and activity of beta1,3-glucuronosyltransferase I (GlcAT-I). Protein Sci. 2006 Jul;15(7):1667-78. Pubmed: 16815917
- Ghosal A, Hapangama N, Yuan Y, Achanfuo-Yeboah J, Iannucci R, Chowdhury S, Alton K, Patrick JE, Zbaida S: Identification of human UDP-glucuronosyltransferase enzyme(s) responsible for the glucuronidation of ezetimibe (Zetia). Drug Metab Dispos. 2004 Mar;32(3):314-20. Pubmed: 14977865
- Hagenauer B, Salamon A, Thalhammer T, Kunert O, Haslinger E, Klingler P, Senderowicz AM, Sausville EA, Jager W: In vitro glucuronidation of the cyclin-dependent kinase inhibitor flavopiridol by rat and human liver microsomes: involvement of UDP-glucuronosyltransferases 1A1 and 1A9. Drug Metab Dispos. 2001 Apr;29(4 Pt 1):407-14. Pubmed: 11259324
- Huskey SW, Doss GA, Miller RR, Schoen WR, Chiu SH: N-glucuronidation reactions. II. Relative N-glucuronidation reactivity of methylbiphenyl tetrazole, methylbiphenyl triazole, and methylbiphenyl imidazole in rat, monkey, and human hepatic microsomes. Drug Metab Dispos. 1994 Jul-Aug;22(4):651-8. Pubmed: 7956743
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Quintus J, Kovar KA, Link P, Hamacher H: Urinary excretion of arbutin metabolites after oral administration of bearberry leaf extracts. Planta Med. 2005 Feb;71(2):147-52. Pubmed: 15729623
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: | Simon, Ethan S.; Grabowski, Sven; Whitesides, George M. Convenient syntheses of cytidine 5'-triphosphate, guanosine 5'-triphosphate, and uridine 5'-triphosphate and their use in the preparation of UDP-glucose, UDP-glucuronic acid, and GDP-mannose. Journal |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|