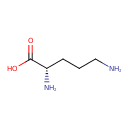
Ornithine (ECMDB00214) (M2MDB000087)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 10:23:49 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-09-13 12:56:07 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Ornithine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Ornithine is an amino acid produced in the urea cycle by the splitting off of urea from arginine. It is a central part of the urea cycle, which allows for the disposal of excess nitrogen. L-Ornithine is also a precursor of citrulline and arginine. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C5H12N2O2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 132.161 Monoisotopic: 132.089877638 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | AHLPHDHHMVZTML-BYPYZUCNSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C5H12N2O2/c6-3-1-2-4(7)5(8)9/h4H,1-3,6-7H2,(H,8,9)/t4-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 70-26-8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2,5-diaminopentanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | ornithine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | NCCC[C@H](N)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | L-alpha-amino acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 140 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + Ornithine > ADP + Hydrogen ion + Ornithine + Phosphate Adenosine triphosphate + Water + Ornithine > ADP + Hydrogen ion + Ornithine + Phosphate Hydrogen ion + Ornithine + L-Ornithine <> Carbon dioxide + Putrescine + Ethylenediamine Carbamoylphosphate + Ornithine + L-Ornithine <> Citrulline + Hydrogen ion + Phosphate N-Acetylornithine + Water <> Acetic acid + Ornithine + L-Ornithine N-Acetylornithine + Water <> Acetic acid + Ornithine Ornithine <> Putrescine + Carbon dioxide Carbamoylphosphate + Ornithine <> Phosphate + Citrulline Adenosine triphosphate + Ornithine + Water > ADP + Phosphate + Ornithine + Hydrogen ion Adenosine triphosphate + Ornithine + Water > ADP + Phosphate + Ornithine + Hydrogen ion N-Acetylornithine + Water > Ornithine + Acetic acid Ornithine + Carbamoylphosphate <> Hydrogen ion + Citrulline + Phosphate Hydrogen ion + Ornithine > Carbon dioxide + Putrescine Ornithine > Putrescine + Carbon dioxide Carbamoylphosphate + Ornithine > Inorganic phosphate + Citrulline N-Acetylornithine + Water > Ornithine + Acetic acid + Ornithine Ornithine + Carbamoylphosphate + Ornithine > Phosphate + Hydrogen ion + Citrulline Ornithine + Hydrogen ion + Ornithine > Putrescine + Carbon dioxide Carbamoylphosphate + Ornithine + L-Ornithine <> Citrulline + Hydrogen ion + Phosphate Hydrogen ion + Ornithine + L-Ornithine <> Carbon dioxide + Putrescine + Ethylenediamine Ornithine <> Putrescine + Carbon dioxide N-Acetylornithine + Water <> Acetic acid + Ornithine + L-Ornithine N-Acetylornithine + Water <> Acetic acid + Ornithine + L-Ornithine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Find out more about how we convert literature concentrations. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Zhang, Peng; Zhang, Shurong; Liu, Chunqiao; Yang, Yuhong. Method for preparing L-ornithine by enzymatic conversion. Faming Zhuanli Shenqing Gongkai Shuomingshu (2007), 8pp. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in carboxyl- or carbamoyltransferase activity
- Specific function:
- Carbamoyl phosphate + L-ornithine = phosphate + L-citrulline
- Gene Name:
- argI
- Uniprot ID:
- P04391
- Molecular weight:
- 36907
Reactions
| Carbamoyl phosphate + L-ornithine = phosphate + L-citrulline. |
- General function:
- Involved in carboxyl- or carbamoyltransferase activity
- Specific function:
- Carbamoyl phosphate + L-ornithine = phosphate + L-citrulline
- Gene Name:
- argF
- Uniprot ID:
- P06960
- Molecular weight:
- 36827
Reactions
| Carbamoyl phosphate + L-ornithine = phosphate + L-citrulline. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the binding-protein-dependent transport system for histidine. Probably responsible for energy coupling to the transport system
- Gene Name:
- hisP
- Uniprot ID:
- P07109
- Molecular weight:
- 28653
- General function:
- Involved in carboxy-lyase activity
- Specific function:
- L-ornithine = putrescine + CO(2)
- Gene Name:
- speC
- Uniprot ID:
- P21169
- Molecular weight:
- 79416
Reactions
| L-ornithine = putrescine + CO(2). |
- General function:
- Involved in zinc ion binding
- Specific function:
- Displays a broad specificity and can also deacylate substrates such as acetylarginine, acetylhistidine or acetylglutamate semialdehyde
- Gene Name:
- argE
- Uniprot ID:
- P23908
- Molecular weight:
- 42347
Reactions
| N(2)-acetyl-L-ornithine + H(2)O = acetate + L-ornithine. |
- General function:
- Involved in carboxy-lyase activity
- Specific function:
- L-ornithine = putrescine + CO(2)
- Gene Name:
- speF
- Uniprot ID:
- P24169
- Molecular weight:
- 82415
Reactions
| L-ornithine = putrescine + CO(2). |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for histidine; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- hisM
- Uniprot ID:
- P0AEU3
- Molecular weight:
- 26870
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for histidine; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- hisQ
- Uniprot ID:
- P52094
- Molecular weight:
- 24649
- General function:
- Involved in transporter activity
- Specific function:
- This periplasmic binding protein is involved in an arginine transport system. ArgT and histidine-binding protein J (hisJ) interact with a common membrane-bound receptor, hisP
- Gene Name:
- argT
- Uniprot ID:
- P09551
- Molecular weight:
- 27991
Transporters
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the binding-protein-dependent transport system for histidine. Probably responsible for energy coupling to the transport system
- Gene Name:
- hisP
- Uniprot ID:
- P07109
- Molecular weight:
- 28653
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for histidine; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- hisM
- Uniprot ID:
- P0AEU3
- Molecular weight:
- 26870
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for histidine; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- hisQ
- Uniprot ID:
- P52094
- Molecular weight:
- 24649
- General function:
- Involved in transporter activity
- Specific function:
- This periplasmic binding protein is involved in an arginine transport system. ArgT and histidine-binding protein J (hisJ) interact with a common membrane-bound receptor, hisP
- Gene Name:
- argT
- Uniprot ID:
- P09551
- Molecular weight:
- 27991
- General function:
- Involved in transporter activity
- Specific function:
- Non-specific porin
- Gene Name:
- ompN
- Uniprot ID:
- P77747
- Molecular weight:
- 41220
- General function:
- Involved in transporter activity
- Specific function:
- Uptake of inorganic phosphate, phosphorylated compounds, and some other negatively charged solutes
- Gene Name:
- phoE
- Uniprot ID:
- P02932
- Molecular weight:
- 38922
- General function:
- Involved in amino acid transmembrane transporter activity
- Specific function:
- Catalyzes an electroneutral exchange between arginine and ornithine to allow high-efficiency energy conversion in the arginine deiminase pathway
- Gene Name:
- ydgI
- Uniprot ID:
- P0AAE5
- Molecular weight:
- 49501
- General function:
- Involved in transporter activity
- Specific function:
- OmpF is a porin that forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane. It is also a receptor for the bacteriophage T2
- Gene Name:
- ompF
- Uniprot ID:
- P02931
- Molecular weight:
- 39333
- General function:
- Involved in amino acid transmembrane transporter activity
- Specific function:
- Probable putrescine-ornithine antiporter
- Gene Name:
- potE
- Uniprot ID:
- P0AAF1
- Molecular weight:
- 46495
- General function:
- Involved in transporter activity
- Specific function:
- Forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane
- Gene Name:
- ompC
- Uniprot ID:
- P06996
- Molecular weight:
- 40368