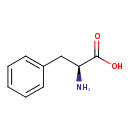
L-Phenylalanine (ECMDB00159) (M2MDB000061)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 10:22:16 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-09-13 12:56:06 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | L-Phenylalanine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Phenylalanine is an essential amino acid and the precursor for the amino acid tyrosine. Phenylalanine is a precursor of the neurotransmitters called catecholamines, which are adrenalin-like substances. Normal metabolism of phenylalanine requires biopterin, iron, niacin, vitamin B6, copper and vitamin C. Phenylalanine and tyrosine, like L-dopa, produce a catecholamine effect. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C9H11NO2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 165.1891 Monoisotopic: 165.078978601 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | COLNVLDHVKWLRT-QMMMGPOBSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C9H11NO2/c10-8(9(11)12)6-7-4-2-1-3-5-7/h1-5,8H,6,10H2,(H,11,12)/t8-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 63-91-2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-amino-3-phenylpropanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | L-phenylalanine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | N[C@@H](CC1=CC=CC=C1)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as phenylalanine and derivatives. Phenylalanine and derivatives are compounds containing phenylalanine or a derivative thereof resulting from reaction of phenylalanine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Phenylalanine and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic homomonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 283 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + L-Phenylalanine + tRNA(Phe) + tRNA(Phe) <> Adenosine monophosphate + L-Phenylalanyl-tRNA(Phe) + Pyrophosphate + L-Phenylalanyl-tRNA(Phe) alpha-Ketoglutarate + L-Phenylalanine <> L-Glutamate + Phenylpyruvic acid L-Arogenate <> L-Phenylalanine + Water + Carbon dioxide L-Phenylalanine + Oxygen <> 2-Phenylacetamide + Carbon dioxide Adenosine triphosphate + L-Phenylalanine + tRNA(Phe) <> Adenosine monophosphate + Pyrophosphate + L-Phenylalanyl-tRNA(Phe) Phenylpyruvic acid + L-Glutamate <> L-Phenylalanine + Oxoglutaric acid Adenosine triphosphate + L-Phenylalanine + tRNA(Phe) > Adenosine monophosphate + Pyrophosphate + L-phenylalanyl-tRNA(Phe) L-Phenylalanine + Adenosine triphosphate + Hydrogen ion + tRNA(Phe) + L-Phenylalanine > Adenosine monophosphate + Pyrophosphate + L-phenylalanyl-tRNA(Phe) Phenylpyruvic acid + L-Glutamic acid + L-Glutamate > Oxoglutaric acid + L-Phenylalanine + L-Phenylalanine L-Phenylalanine + Oxygen + L-Phenylalanine <> Oxoglutaric acid + Phenylpyruvic acid L-Phenylalanine + Oxygen + L-Phenylalanine <> Carbon dioxide + Sinapyl alcohol Adenosine triphosphate + L-Phenylalanine + tRNA(Phe) <> Adenosine monophosphate + L-Phenylalanyl-tRNA(Phe) + Pyrophosphate alpha-Ketoglutarate + L-Phenylalanine <> L-Glutamate + Phenylpyruvic acid Adenosine triphosphate + L-Phenylalanine + tRNA(Phe) <> Adenosine monophosphate + L-Phenylalanyl-tRNA(Phe) + Pyrophosphate alpha-Ketoglutarate + L-Phenylalanine <> L-Glutamate + Phenylpyruvic acid alpha-Ketoglutarate + L-Phenylalanine <> L-Glutamate + Phenylpyruvic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Find out more about how we convert literature concentrations. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Zhou, Hua; Zhong, Yao; Sun, Guanghai; Wei, Ping. Preparation of L-phenylalanine by an aqueous two-phase system. Huaxue Fanying Gongcheng Yu Gongyi (2006), 22(2), 146-150. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in transferase activity
- Specific function:
- L-aspartate + 2-oxoglutarate = oxaloacetate + L-glutamate
- Gene Name:
- aspC
- Uniprot ID:
- P00509
- Molecular weight:
- 43573
Reactions
| L-aspartate + 2-oxoglutarate = oxaloacetate + L-glutamate. |
- General function:
- Involved in transferase activity
- Specific function:
- An aromatic amino acid + 2-oxoglutarate = an aromatic oxo acid + L-glutamate
- Gene Name:
- tyrB
- Uniprot ID:
- P04693
- Molecular weight:
- 43537
Reactions
| An aromatic amino acid + 2-oxoglutarate = an aromatic oxo acid + L-glutamate. |
- General function:
- Involved in transferase activity
- Specific function:
- L-histidinol phosphate + 2-oxoglutarate = 3- (imidazol-4-yl)-2-oxopropyl phosphate + L-glutamate
- Gene Name:
- hisC
- Uniprot ID:
- P06986
- Molecular weight:
- 39360
Reactions
| L-histidinol phosphate + 2-oxoglutarate = 3-(imidazol-4-yl)-2-oxopropyl phosphate + L-glutamate. |
- General function:
- Involved in RNA binding
- Specific function:
- ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe)
- Gene Name:
- pheT
- Uniprot ID:
- P07395
- Molecular weight:
- 87377
Reactions
| ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe). |
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe)
- Gene Name:
- pheS
- Uniprot ID:
- P08312
- Molecular weight:
- 36832
Reactions
| ATP + L-phenylalanine + tRNA(Phe) = AMP + diphosphate + L-phenylalanyl-tRNA(Phe). |
- General function:
- Involved in chorismate mutase activity
- Specific function:
- Chorismate = prephenate
- Gene Name:
- pheA
- Uniprot ID:
- P0A9J8
- Molecular weight:
- 43111
Reactions
| Chorismate = prephenate. |
| Prephenate = phenylpyruvate + H(2)O + CO(2). |
- General function:
- Involved in catalase activity
- Specific function:
- Bifunctional enzyme with both catalase and broad- spectrum peroxidase activity. Displays also NADH oxidase, INH lyase and isonicotinoyl-NAD synthase activity
- Gene Name:
- katG
- Uniprot ID:
- P13029
- Molecular weight:
- 80023
Reactions
| Donor + H(2)O(2) = oxidized donor + 2 H(2)O. |
| 2 H(2)O(2) = O(2) + 2 H(2)O. |
- General function:
- Involved in catalytic activity
- Specific function:
- Acts on leucine, isoleucine and valine
- Gene Name:
- ilvE
- Uniprot ID:
- P0AB80
- Molecular weight:
- 34093
Reactions
| L-leucine + 2-oxoglutarate = 4-methyl-2-oxopentanoate + L-glutamate. |
| L-isoleucine + 2-oxoglutarate = (S)-3-methyl-2-oxopentanoate + L-glutamate. |
| L-valine + 2-oxoglutarate = 3-methyl-2-oxobutanoate + L-glutamate. |
Transporters
- General function:
- Involved in nucleotide binding
- Specific function:
- Probably part of a binding-protein-dependent transport system yecCS for an amino acid. Probably responsible for energy coupling to the transport system
- Gene Name:
- yecC
- Uniprot ID:
- P37774
- Molecular weight:
- 27677
- General function:
- Involved in transporter activity
- Specific function:
- Probably part of the binding-protein-dependent transport system yecCS for an amino acid; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- yecS
- Uniprot ID:
- P0AFT2
- Molecular weight:
- 24801
- General function:
- Involved in transport
- Specific function:
- Permease that is involved in the transport across the cytoplasmic membrane of the aromatic amino acids (phenylalanine, tyrosine, and tryptophan)
- Gene Name:
- aroP
- Uniprot ID:
- P15993
- Molecular weight:
- 49690
- General function:
- Involved in transport
- Specific function:
- Permease that is involved in the transport across the cytoplasmic membrane of phenylalanine
- Gene Name:
- pheP
- Uniprot ID:
- P24207
- Molecular weight:
- 50677
- General function:
- Involved in transporter activity
- Specific function:
- Non-specific porin
- Gene Name:
- ompN
- Uniprot ID:
- P77747
- Molecular weight:
- 41220
- General function:
- Carbohydrate transport and metabolism
- Specific function:
- Specific function unknown
- Gene Name:
- yddG
- Uniprot ID:
- P46136
- Molecular weight:
- 31539
- General function:
- Involved in transporter activity
- Specific function:
- Uptake of inorganic phosphate, phosphorylated compounds, and some other negatively charged solutes
- Gene Name:
- phoE
- Uniprot ID:
- P02932
- Molecular weight:
- 38922
- General function:
- Involved in transporter activity
- Specific function:
- OmpF is a porin that forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane. It is also a receptor for the bacteriophage T2
- Gene Name:
- ompF
- Uniprot ID:
- P02931
- Molecular weight:
- 39333
- General function:
- Involved in transporter activity
- Specific function:
- Forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane
- Gene Name:
- ompC
- Uniprot ID:
- P06996
- Molecular weight:
- 40368